resDNASEQ® E. coli Residual DNA Quantitation System
Posted: 1 June 2012 | | No comments yet
Integrated sample preparation and real-time PCR assay for the quantitation of E. coli host cell DNA…

resDNASEQ Kit
Integrated sample preparation and real-time PCR assay for the quantitation of E. coli host cell DNA
- Highly sensitive quantitation using proven TaqMan® real-time qPCR technology (Figure 1)
- Manual and automated sample preparation, optimized for quantitative recovery from complex sample matrices (Table 1)
- Enables consistent performance across the expected range of DNA fragment sizes (Figure 2)
- Integrated system from sample to results with sample preparation, master mix, TaqMan® primer/probe mix, and genomic DNA standard


resDNASEQ Kit
The resDNASEQ® E. coli Residual DNA Quantitation System is a quantitative PCR (qPCR)-based system for the detection of host cell DNA from E. coli, an expression system commonly used for the production of recombinant proteins. Reliable and rapid, the system enables sensitive (LOQ = 15 pg DNA/mL test sample, Figure 1) and specific (Figures 2 and 3) quantitation of E. coli DNA typically in less than four hours. This performance helps ensure a high degree of confidence in quantitation data obtained from a broad range of sample types—from in-process samples to bulk drug substance—whether the sample contains high molecular weight or sheared DNA (Figure 3).
Table 1. DNA recovery using the manual PrepSEQ® sample preparation protocol. Assay performance data using 10 pg E. coli genomic DNA spike per sample, 3 analysts, and 9 test samples.
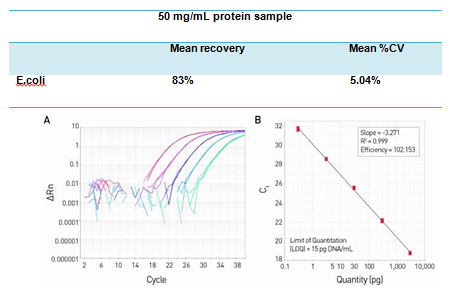

Figure 1. High sensitivity and broad dynamic range using the resDNASEQ® E. coli Residual DNA Quantitation System. The amplification plot (A) was generated using a 10-fold serial dilution of E. coli genomic DNA, provided in the kit. Concentrations range from 3 ng to 300 fg. The standard curve (B) of the 10-fold dilution series. Data were analyzed using AccuSEQ® Real-Time Detection Software.
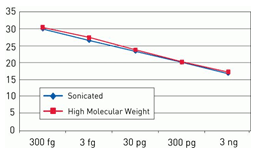

Figure 2. Consistent quantitation across a broad range of fragment sizes. Standard curves were generated using a 10-fold serial dilution of high molecular weight (red) and fragmented (blue) DNA from 3 ng to 300 fg. Fragmented DNA was generated by sonicating total E. coli genomic DNA. Fragmentation of the DNA was confirmed by agarose gel analysis.
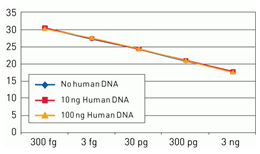

Figure 3. Assay specificity. Standard curves generated using 10-fold serial dilution (3 ng to 300 fg) of E. coli genomic DNA (included in the kit) in the presence of 100 ng human DNA (yellow), 10 ng human DNA (red), and no human DNA (blue).
To learn more, please visit: https://products.invitrogen.com/ivgn/product/4460366?ICID=search-product