Real-time PCR: clinical applications
Posted: 24 March 2006 | | No comments yet
The real-time PCR technique is one of the emerging techniques that, although only described for the first time about a decade ago, have become the method of choice for quantification of DNA and RNA levels in cells, tissues and tissue biopsies.
The real-time PCR technique is one of the emerging techniques that, although only described for the first time about a decade ago, have become the method of choice for quantification of DNA and RNA levels in cells, tissues and tissue biopsies.
The real-time PCR technique is one of the emerging techniques that, although only described for the first time about a decade ago, have become the method of choice for quantification of DNA and RNA levels in cells, tissues and tissue biopsies.
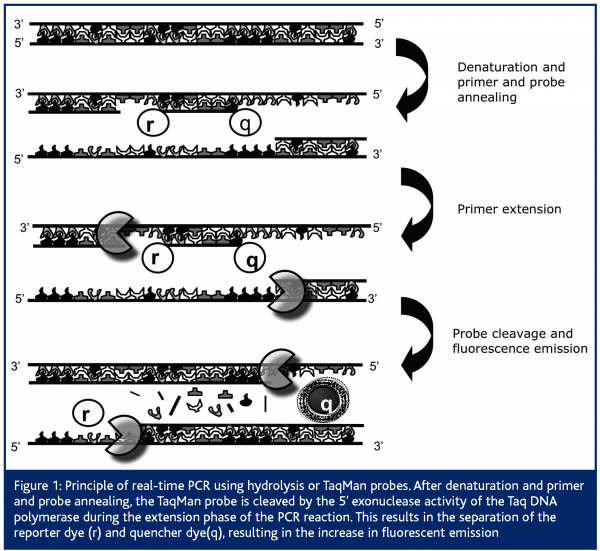
Comparable to classical end-point PCR, it is based on the amplification of DNA by the Thermus aquaticus (Taq) polymerase. Apart from this, the real-time PCR technique also makes use of the 5’-3’ exonuclease activity of this same Taq polymerase to cleave a dual-labeled fluorescent probe, giving rise to fluorescent emission1,2. Therefore, next to the presence of two primers in the PCR reaction, real-time PCR makes use of an additional oligonucleotide primer that is dually labeled with two fluorophores. Importantly, this oligonucleotide primer, also called hydrolysis or TaqMan probe, anneals to the target internally of the two primers and emits a fluorescent signal only upon cleavage. In its free, intact form no fluorescent emission can be measured because fluorescent emission of the reporter dye is absorbed by the quenching dye. Quantification of the initial amount of target DNA is based on the increase in fluorescent emission, measured in every cycle of the PCR reaction1,2. Therefore, PCR amplification and data acquisition (by measuring the increase in fluorescent emission as the PCR reaction proceeds) are performed in one single step, such that no post-PCR processing is required (Figure 1). Next to this classical specific detection chemistry, other specific probe systems have been developed, which have been discussed in a previously published review3.
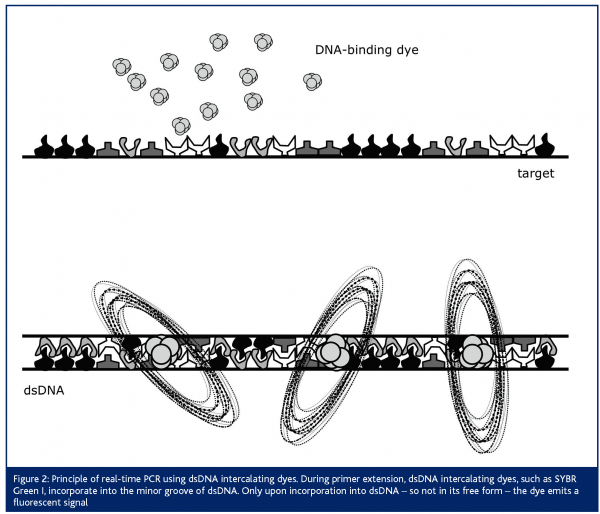
Next to the use of this so-called ‘TaqMan’ probe, real-time PCR assays have been developed, making use of a dsDNA-intercalating dye4,5 (Figure 2). For this purpose, SYBR Green I is most often applied. This dye does not emit fluorescent light in its free form, but only upon incorporation into dsDNA. An advantage when choosing this strategy is the lower running cost of the assay, since no expensive dual-labeled probe is necessary. An important disadvantage however, is the lower specificity of the assay. Evidently, when measuring the fluorescent emission during PCR amplification, aspecific amplification products or primer-dimers that may be formed will also be measured. Therefore, careful optimisation of this assay is crucial for obtaining accurate results. One way to control these aspecific amplification or primer-dimer formation is by performing a melting-curve analysis, subsequent to the PCR reaction. Principally, this analysis is based on the knowledge that each PCR product will have a specific melting temperature, while aspecific amplification products or primer-dimers (with a different amplicon length and different %GC) will have another melting temperature. The new-generation real-time PCR instruments are all able to perform a melting-curve analysis. Thus, although optimisation of the assay is more time-consuming, one can control for this potential problem.
Since its invention in the mid 1990s, real-time PCR has evolved into a research tool that is present on almost every laboratory bench. Its widespread use has been facilitated by different cooperating factors: 1) PCR amplification and detection are performed in one single step, which significantly reduces hands-on-time (no post-PCR analysis, such as, for example, separation on agarose gels, is required) as well as contamination (PCR tubes do not have to be opened after amplification); 2) a wide dynamic range makes it possible to quantify DNA/RNA samples of highly different abundance within a single experiment; 3) analysis can be performed on tiny amounts of tissue or tissue biopsies; 4) reproducibility of the method is extremely high. Moreover, the use of the technique has been greatly facilitated for high throughput use by the introduction of robotic workstations for automatic DNA/RNA extraction and preparation of PCR reactions. In addition, the availability of a broad variety of real-time PCR instrumentation makes it possible to purchase a suitable instrument for each ones specific needs, be it for instance RNA quantification or pathogen detection. Finally, due to the development of a whole range of different PCR kits and alternative chemistries and the natural competition between different commercial suppliers the technique has become affordable for routine analysis in almost every clinical and molecular laboratory.
Quantification of gene expression by real-time RT-PCR
Undoubtedly, the most widely used application of real-time PCR is the quantification of mRNA expression, or real-time reverse-transcriptase PCR (RT-PCR). When optimising this technique different aspects need special attention, among which 1) high RNA quality, 2) normalisation and choice of a suitable housekeeping gene, 3) optimal assay design and template preparation and 4) optimal data analysis and interpretation of the results3.
Although used in all fields of fundamental research and clinical diagnostics one can think of, an excellent example where real-time RT-PCR has become the ‘gold standard’ for routine analysis is the quantification of cytokine and chemokine gene expression. Cytokines are a large group of low molecular weight proteins, secreted by many different cell types, which play a central role in modulating immune responses. As a result, abnormal cytokine expression levels will contribute to immune-mediated disorders, such as autoimmune, allergic and inflammatory diseases or to transplant rejection. Chemokines are a subgroup of cytokines which function as chemo-attractants, controlling the trafficking of specific subsets of leukocytes to sites of tissue damage. In autoimmune diseases, such as for instance type 1 diabetes, the migration and accumulation of immune cells to diseased target organs is a critical step in the development of the disease.
It is clear that a reliable mRNA quantification of cytokines, chemokines and their receptors in diseased target organs are fundamental to our understanding of different inflammatory diseases and for monitoring therapeutic effects. Different aspects have contributed to the fact that real-time RT-PCR quantification for analysis of cytokines and chemokines has found such an exponential increase in use, such as the availability of only tiny amounts of tissue or tissue biopsies and the often low expression level of these immune mediators. Moreover, optimised and validated primer and TaqMan probes have been published for a large panel of cytokines and chemokines6,7, or can be purchased commercially. The widespread use of their accurate and sensitive quantification definitely refined the role of cytokines and chemokines in numerous biological and clinical contexts.
Clinical applications
The use of real time PCR for a number of clinical applications has increased substantially. For instance in the field of clinical microbiology, oncology and gene therapy it is clear that it has become the standard method for disease detection. Specific factors have to be taken into account when working with clinical samples, which do not apply to assays optimised for fundamental molecular research. Of major importance is a standardisation of the test across different laboratories. Also relevant positive and negative controls need to be included in all assays. Furthermore, optimal RNA quality is of utmost importance in order to obtain reliable quantitative PCR results8,9.
Clinical microbiology
Real-time PCR has been shown to be extremely useful for quantification of viruses and bacteria, and to a lesser extent also for fungi, parasites and protozoans10. Most of the assays described in literature allow an increased sensitivity and enhanced speed of microbial detection as compared to the classically used culture techniques. Moreover the assay makes an accurate quantification of pathogen load possible, opposed to previously obtained qualitative data. An aspect that must be taken into account when designing real-time assays for pathogen detection is that most infectious agents are characterised by a high mutation rate, which can dramatically influence the pathogen load estimation. This can be overcome by designing primers in highly conserved regions. Alternatively, of course, sequence variations can provide the basis for development of subtype-specific assays.
From the countless number of publications that have used real-time PCR for the detection of viral load, it is obvious that this technique has proved its usefulness as an indicator of the extent of active infection, interactions between virus and host, and the changes in viral DNA copy numbers as a result of anti-viral therapy11. All these factors play an important role in the treatment regimen envisaged.
Also for the detection of bacterial infections, the real-time PCR technique has been a great step forward in terms of effective antibiotic treatment. Indeed, because of the rapid and accurate real-time detection, the infection status of the patient is much clearer for the clinician, allowing a more specific and timely application of antibiotic treatment.
Another application in this area is the quantification of gene transfer vectors used for gene therapy. Considering gene therapy as a drug delivery system, two important parameters have to be analysed: 1) the expression level of the therapeutic gene and 2) the distribution of the drug in different organs. For the control of both of these parameters, real-time PCR assays have been validated, especially concentrating on the accuracy and sensitivity of the assay12.
Clinical oncology
In the field of clinical oncology major efforts have been undertaken to optimise real-time PCR techniques for the detection of minimal residual disease, single nucleotide polymorphisms and chromosomal translocations.
In many hematological malignancies the detection of minimal residual disease (MRD) directly correlates with the clinical outcome of the disease. Therefore, MRD monitoring is important for therapy guidance in clinical settings. Real-time PCR based techniques for the detection of MRD are used in clinical protocols. The major challenge when optimising such assays is to develop standardised protocols between different laboratories. Importantly, a large scale collaboration effort has been undertaken, within the Europe Against Cancer (EAC) program, to establish a standardised protocol for real-time PCR of the main leukemia-associated fusion gene measurements, that can be applied in 35-45 per cent of acute lymphoblastic leukemia and acute myeloid leukemia and in 90 per cent of chronic myeloid leukemia8. This collaboration included the design and selection of optimal primer/probe sets, choice of optimal standard curves, standardisation of the protocols, performance of quality control rounds and choice of universal normalisation genes.
Single nucleotide polymorphisms (SNPs) present in the human genome are a powerful tool in the study of genetic factors associated with human diseases. When optimising real-time PCR assays for this application the level of discrimination between target and non-target allele is of crucial importance. Most often, one set of primers and two allele-specific fluorescent labeled probes are used, making use of different reporter dyes. The two alleles can thus be distinguished by the differential fluorescent emission of the two different reporter dyes. Alternatively, SNP detection is often making use of molecular beacons or scorpion probes. More recently, new probe designs, such as peptide nucleic acid (PNA), locked nucleic acid (LNA) or minor groove binding (MGB) probes have been evaluated for this purpose. These probes result in a shorter probe design with higher sequence-specific detection, making discrimination between a single nucleotide mismatches easier13.
Chromosomal translocations, which are often taking place in tumour cells, can be employed as tumour-specific PCR targets. These fusion genes are directly related to the oncogenic process and therefore stable throughout the disease course. Most often, for the design of such assays, the primers are chosen to anneal to opposite sides of the breakpoint in the fusion gene. A major challenge is the relatively large amplicon size often necessary to span the fusion breakpoints, especially if one wants to design an assay for different patients clustering in relatively small breakpoint areas.
Conclusions
Real-time PCR has greatly simplified the quantification of DNA and RNA. This has had a great impact in the field of molecular research and clinical diagnostics, since enormous amounts of data can be obtained within a very short research time. The decreased costs for real-time thermal cyclers and reagents for applying the technique, as well as the development of high throughput robotic systems for preparation of the reactions, have aided its rapid increase in use. Therefore, it seems that this technique is becoming the gold standard for the detection and quantification of DNA and RNA.
Although the real-time PCR technique is very accurate, sensitive and reproducible, the reliability of the data is largely dependent on several other factors. These factors, such as sample preparation, quality of the RNA, choice of a housekeeping gene and normalisation of samples, need careful consideration and optimisation. Furthermore, in a clinical setting it is important that standardised criteria and international uniformity in experimental design and data analysis is reached, in order to be able to compare data between different clinical laboratories. Great efforts are being undertaken to overcome these difficulties, resulting in a further improvement of the technique, which will definitely aid to its more widespread use.


References
- Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 6:986-94, 1996.
- Gibson UE, Heid CA, Williams PM. A novel method for real time quantitative RT-PCR. Genome Res. 6:995-1001, 1996.
- Giulietti A, Overbergh L, Valckx D, Decallonne B, Bouillon R, Mathieu C. An overview of real-time quantitative PCR: applications to quantify cytokine gene expression. Review. Methods 25:386-401. 2001.
- Higuchi R, Fockler C, Dollinger G, Watson R. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology 11:1026-30, 1993.
- Ririe KM, Rasmussen RP, Wittwer CT. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal Biochem. 245:154-60, 1997.
- Overbergh L, Giulietti A, Valckx D, Decallonne R, Bouillon R, Mathieu C., The use of real-time reverse transcriptase PCR for the quantification of cytokine gene expression. J Biomol Tech. 14:33-43, 2003.
- Overbergh L, Gysemans C and Mathieu C. Quantification of chemokines by real-time RT-PCR: Applications in type 1 diabetes. Expert Rev Mol Diagn. In press, 2006.
- Gabert J, Beillard E, van der Velden VHJ, Bi W, Grimwade D, Pallisgaard N, Barbany G, Cazzaniga G, Cayuela JM, Cavé H, Pane F, Aerts JLE, De Micheli D, Thirion X, Prdal V, Gonzalez M, Viehmann S, Malec M, Saglio G and van Dongen JJM. Standardization and quality control studies of ‘real-time’ quantitative reverse transcriptase polymerase chain reaction of fusion gene transcripts for residual disease detection in leukemia – A Europe Against Cancer Program. Leukemia 17:2318-2357, 2003.
- Van der Velden VHJ. Differential stability of control gene and fusion gene transcripts over time may hamper accurate quantification of minimal residual disease – a study within the Eurpoe Against Cancer Program. Leukemia 18:884-886, 2004.
- Mackay IM. Real-time PCR in the microbiology laboratory. Review. Clin Microbiol Infect 10:190-212, 2004.
- Niesters HGM. Molecular and diagnostic clinical virology in real time. Clin Microbiol Infect 10:5-11, 2004.
- Wang F, Puddy AC, Mathis BC, Montalvo AG, Louis AA, McMackin JL, Xu J, Zhang Y, Tan CY, Schofield TL, Wolf JJ and Lewis JA. Using QPCR to assign infectious potencies to adenovirus based vaccines and vectors for gene therapy: toward a universal method for the facile quantitation of virus and vector potency. Vaccine 23: 4500-4508, 2005.
- Gibson NJ. The use of real-time PCR methods in DNA sequence variation. Clin Chim Acta 363:32-47, 2006.