The HUPO Brain Proteome Project
Posted: 2 February 2006 | | No comments yet
The proteome analysis started by the Human Proteome Organization (HUPO)1 is the second big international consortium project after the sequencing of the human genome by the Human Genome Project (HUGO)2. The aim of the HUPO Brain Proteome Project (BPP)3 is to derive in depth knowledge of the brain from analysing samples with state-of-the-art proteomics techniques.

The proteome analysis started by the Human Proteome Organization (HUPO)1 is the second big international consortium project after the sequencing of the human genome by the Human Genome Project (HUGO)2. The aim of the HUPO Brain Proteome Project (BPP)3 is to derive in depth knowledge of the brain from analysing samples with state-of-the-art proteomics techniques.
Two pilot studies have been started to differentially compare human and mouse brain samples that have been analysed in laboratories worldwide. The participating labs received both autopsy and biopsy brain samples for the human pilot study and samples of three different age stages have been sent for investigation in the mouse pilot study. Besides the differential gel and mass spectrometry analysis, complementary methods such as mRNA and Peptidomic analysis will be applied to give a broader insight into the brain constitution.
Proteomics studies driven by large consortia often lead to heterogeneous data due to different strategies, techniques and equipment. To assure a common, standardised interpretation, relevant experimental data should be collected in one database. A suitable database concept has to be defined from the very beginning to avoid technical pitfalls and extensive redesign at a later stage.
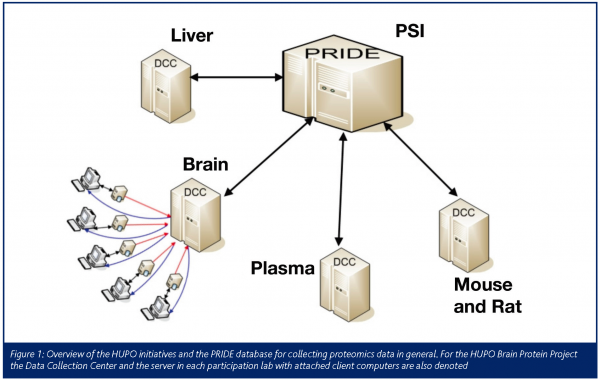
For that purpose the HUPO BPP established a Data Collection Centre (DCC) for storing gathered data and information gained from the experiments4. To produce reliable, reproducible and comparable results the Bioinformatics Committee agreed to perform a reprocessing of all collected data5 The details of execution adhere to the ‘DCC Data Reprocessing Guideline’ which has been published and discussed online (www.hbpp.org). The reprocessing will bring the heterogeneous data to a precisely defined stage from which further data analyses will start.
The heterogeneity of the data due to the application of different methods and use of diverse mass spectrometers are increasing the need for standardisation. To compare these heterogenic data it is important to determine the right procedure to unify the results and to distinguish between false and true positives. Here we describe the approach of analysing heterogeneous data by four different search engines.
For in depth analyses with different scientific topics dedicated task forces have been built. The 2D-gel-images task force will do further analyses of the raw images and re-projection to the original gels6. The results will be correlated back to original MS data and differential information. The raw-data-reprocessing task force has been started by the reanalysis of unprocessed MS data. mRNA profiling; the correlation of the mapped differential expressed gene products with the corresponding proteins; as well as peptidomic interpretation will also be accomplished. Further, deeper insight into the brain proteins will provide analyses such as Gene Ontology, InterPro, disease association, tissue expression, alternative splicing, transmembrane proteins, sorting signals, protein-protein-interaction, pathways and text mining. These will be solely based on the reprocessed protein lists.
After completion of the data analyses phase all applicable data will be provided to the scientific community via the PRIDE repository7 (www.ebi.ac.uk/pride) (Figure 1) at the European Bioinformatics Institute (EBI), Hinxton, UK.
Heterogeneity of experimental data
The heterogeneity of the data is very high owing to the use of diverse analysis strategies and instruments. In total more than one million mass spectra have been submitted to the DCC.
The spectra can be classified in different ways to observe the diversity of experimental setups. Prior to the MS analysis different separation techniques were applied: 32% of the spectra were acquired after 1D gel techniques, 22% after 2D gels and 46% after liquid chromatography. Of the collected spectral data, 82% was produced from human samples. The rest (18%) was generated from mouse samples. The majority of spectra result from MS/MS experiments (99.5%). The remaining 0.5% are MS spectra.
The distribution of mass spectrometric devices is also very heterogenic. Most of the major MS instrument vendors were present with a variety of instruments in one or more labs.
Bioinformatics strategy
In proteomics research the amount of data has increased tremendously in recent years. This increase is due to both the large number of experiments needed to gain significant and statistically sound results and the fact that the number of (for example) mass spectrometric data sets per experiment has increased. Correspondingly the number of software tools (most of which come with their proprietary data formats) has increased as well.
To manage these problems in the pilot study phase of the HUPO BPP, the Bioinformatics Committee decided to store all data in one central database, which is capable of handling the heterogeneous data from different sources.
The DCC is implemented as a two layer client/server architecture based on the proteomics project management software ProteinScapeTM, a development of Bruker Daltonik GmbH and Protagen AG. Most of the participating labs are using ProteinScape locally as a platform to manage their proteomics workflow. All data (i.e. sample descriptions, MS spectra and gel images) is collected via the workplace client software and is sent to the local ProteinScape server, where a first processing will be performed according to the particular expertise of the lab scientists.
After local approval the whole project data can be exported into compressed chunks of 650 MB using a ProteinScape integrated tool and transferred via FTP or mail to the central ProteinScape database at the DCC, which is located at the Medical Proteom-Center (MPC, Bochum, Germany). The common underlying database scheme of ProteinScape for labs and DCC ensures the highest grade of data compatibility and excludes operational dependencies.
The HUPO BPP is one of the first major projects to support the mzData standard of the HUPO Proteomics Standards Initiative (PSI) (psidev.sf.net). Thus, the data submission from the DCC to the PRIDE database, which also gathers data from the other HUPO initiatives and data retrieval from the DCC, will be in an open and standardised way. This will allow upcoming software tools to rapidly gain access to the data gathered by efforts of the HUPO Brain Proteome Project.
Reprocessing
The participating groups have sent their data to the DCC where the central spectra reprocessing was performed.
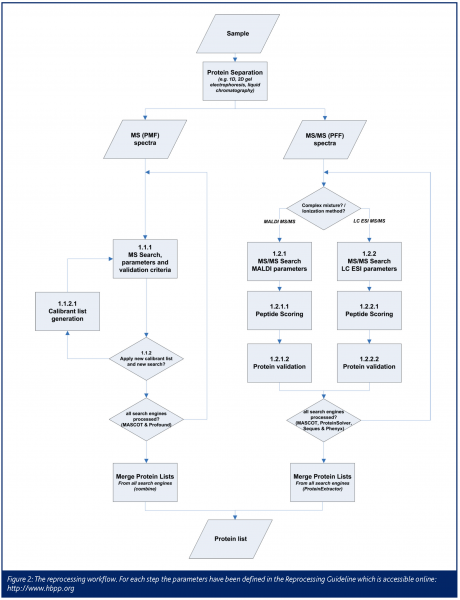
To get the most accurate and reliable information from the gathered data, different protein search engines (Mascot8, Sequest9, Profound, Protein-Solver and Phenyx10) are used, multiplying the amount of processing involved in identifying ed spectra. Matrix Sciences has freely provided additional Mascot licenses for the pilot phase. Genebio also generously provided licenses free of charge for the Phenyx search engine, for all 128 CPUs of the Linux cluster of the Medical Proteom-Center (MPC) in Bochum, Germany where all reprocessing has been done.
To generalise the reprocessing of the diverse data sets, a guideline (see forum.hbpp.org) has been set up defining and standardising all relevant parameters as well as the workflow of protein identification (Figure 1). All different techniques, spectrum types and mass spectrometers are taken into account. Each data set, e.g. from one 2D gel spot, is searched with each of the appropriate search engines to get a peptide identification.
As it is not adequate to use only one parameter set for all analyses, a more flexible way must be applied to assemble protein lists. The peptide identifications that score above a certain threshold are therefore used to generate protein lists by the ProteinExtractor software, which is part of ProteinScape.
All MS data sets are searched against a specially prepared decoy protein database of the International Protein Index (IPI)11 databases for each analysed species. In this decoy database, for each protein of the original database a decoy protein has been added, where all amino acids of the original protein have been shuffled to random positions. The generation of the decoy database has been performed by the decoy database builder, part of the Peakardt software suite (www.peakardt.org). If a search engine claims to have found a peptide that originates from the decoy part of the database it can be assumed that this is a false positive hit. If only the best scoring identified proteins with a fraction of only 5% of decoy peptides are taken as search results, the use of decoy databases will help to assure high quality standards on the identifications. The combination of different search engines with a decoy database strategy will take advantage of each search engine’s specific strengths and guarantees a minimal false positive rate.
From each sample in every lab a protein list is derived. All protein lists are then put together for the final list of human and mouse brain proteins.
Summary
The DCC has been designed to integrate proteomics data (sample information, 1D and 2D gel electrophoresis, mass spectrometry etc.) from participating laboratories with their heterogeneous analysis strategies. It has been successfully set up, revealing its functionality in the international pilot study of the HUPO BPP. The data reprocessing allows an independent reanalysis under standardised criteria.
To gain the maximum amount of information from the heterogeneous data sets different search engines will be used in parallel, utilising the specific strengths of each engine. The estimation of the false positive rate of the protein identifications via a decoy database will assure reliable and high quality results. The parameter settings determined by the false positive rate are used to dynamically adjust the process of generating protein lists.
Outlook
From January 9-11 2006 a jamboree took place at the EBI in Hinxton, U.K. The HUPO BPP Bioinformatics Committee and experts from different fields came together in order to analyse and discuss the reprocessed data. Further results were derived from the submitted data gathered in the course of the pilot phase.
These results will be further discussed at the 5th HUPO BPP Workshop taking place in Dublin, Ireland on February 15 and 16 2006 and will mark the transfer of the HUPO BPP pilot phase into the master phase.
Acknowledgement
The HUPO BPP is supported by the German Ministry of Education and Research (BMBF) with funding 0313318B.
Lennart Martens is a Research Assistant of the Fund for Scientific Research – Flanders (Belgium) (F.W.O. – Vlaanderen).


References
- Hanash, S., HUPO initiatives relevant to clinical proteomics. Mol Cell Proteomics, 2004. 3(4): p. 298-301.
- Lander, E.S., et al., Initial sequencing and analysis of the human genome. Nature, 2001. 409(6822): p. 860-921.
- Meyer, H.E., J. Klose, and M. Hamacher, HBPP and the pursuit of standardisation. Lancet Neurol, 2003. 2(11): p. 657-8.
- Stephan, C., et al., 5th HUPO BPP Bioinformatics Meeting at the European Bioinformatics Institute in Hinxton, UK–Setting the analysis frame. Proteomics, 2005. 5(14): p. 3560-2.
- Stephan, C., et al., HUPO Brain Proteome Project Pilot Studies: bioinformatics at work. Proteomics, 2005. 5(11): p. 2716-7.
- Dowsey, A.W., M.J. Dunn, and G.Z. Yang, ProteomeGRID: towards a high-throughput proteomics pipeline through opportunistic cluster image computing for two-dimensional gel electrophoresis. Proteomics, 2004. 4(12): p. 3800-12.
- Martens, L., et al., PRIDE: the proteomics identifications database. Proteomics, 2005. 5(13): p. 3537-45.
- Perkins, D.N., et al., Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis, 1999. 20(18): p. 3551-67.
- Eng JK, M.A., and Yates JR 3rd, An Approach to Correlate Tandem Mass Spectral Data of Peptides with Amino Acid Sequences in a Protein Database. J Am Soc Mass Spectrom, 1994(5): p. 976-989.
- Colinge, J., et al., High-performance peptide identification by tandem mass spectrometry allows reliable automatic data processing in proteomics. Proteomics, 2004. 4(7): p. 1977-84.
- Kersey, P.J., et al., The International Protein Index: an integrated database for proteomics experiments. Proteomics, 2004. 4(7): p. 1985-8.
Issue
Related topics
Related organisations
Bruker, European Bioinformatics Institute (EBI), Ghent University



