A new approach to ion channel hit identification
Posted: 11 November 2005 | | No comments yet
Advances in assay technology are driving an exciting new era in ion channel drug discovery. Here, we present a case study describing the use of computational chemistry and focussed library screening to drive rapid hit identification.
Advances in assay technology are driving an exciting new era in ion channel drug discovery. Here, we present a case study describing the use of computational chemistry and focussed library screening to drive rapid hit identification.
Advances in assay technology are driving an exciting new era in ion channel drug discovery. Here, we present a case study describing the use of computational chemistry and focussed library screening to drive rapid hit identification.
Ion channels are established targets for drug discovery with approximately 130 drugs marketed to date. Examples include voltage-gated sodium channel blockers for the treatment of arrhythmia and local anaesthesia; calcium channel blockers for treatment of angina and hypertension and ATP-sensitive potassium channel blockers for type II diabetes. In 2003, global sales of ion channel modulators exceeded $6 billion.
Despite this apparent tractability, it is interesting to note that the major chemical classes of ion channel modulators were identified through serendipity. Furthermore, most classes have been in clinical use for many years, pre-dating major milestones in ion channel research such as the development of patch clamp electrophysiology (the gold standard assay format) and molecular cloning of ion channel families. The development of benzodiazepines provides a good example: Chlordiazepoxide was synthesised in 1954 by the late Dr. Leo Sternbach1, working for Hoffmann-La Roche. Although the compound sat on a bench for 18 months, in vivo testing in mice revealed that chlordiazepoxide had significant biological activity, including hypnotic and sedative effects. The compound was launched as Librium in 1960 for the treatment of anxiety and a host of other benzodiazepine drugs followed. It was not until the late 1970s that GABAA receptors were implicated as the biological target of benzodiazepines and a further ten years before GABAA receptor sub-units were cloned and then analysed by electrophysiology. It is now known that the benzodiazepines have an allosteric action on the GABAA channel, prolonging opening following GABA binding2.
Until very recently more targeted ion channel drug discovery, using recombinantly expressed ion channel sub-units, was severely hampered by issues with tractability. Thus, while high-throughput screens were possible using radioligand-binding or ion flux methods, hit validation relied upon the use of the technically demanding and low throughput whole-cell patch clamp technique. With a maximum throughput of around 50 data points per day, conventional patch clamp electrophysiology created a significant bottleneck in the progression path. A further issue arises when comparing IC50 data from patch-clamp assays with values generated in the primary screening format. In many cases, there is a relatively poor correlation, thereby creating an additional problem in that secondary screening during lead identification and optimisation phases is dependent upon the same indirect assay formats that were used for HTS.
As reviewed recently in this journal3, ion channel drug discovery has been revolutionised by the introduction of automated electrophysiology technologies. Thus, for a scientist interested in making increased throughput electrophysiological recording from mammalian cells, eight products from five different manufacturers are now commercially available. Each platform has its own advantages and disadvantages in terms of throughput, applicability to assaying different ion channel sub-families and the level of ion channel expertise required for operation.
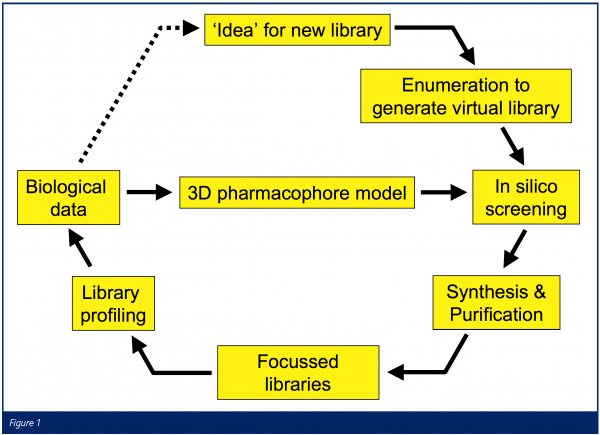
In AstraZeneca, we believe that the combination of a rich pharmacological heritage and advances in screening technology provide an opportunity to apply a more rational approach to ion channel drug design. We have therefore established a process whereby extensive in silico screening of virtual chemical libraries against computational pharmacophore models, followed by multiple parallel synthesis (MPS) chemistry, is used to rapidly generate focussed chemical libraries. Library compounds are then tested using high-throughput electrophysiology methods and the resulting data is used to refine the pharmacophore models and to drive the identification of hit series (Figure 1).
To provide an example, there follows a description of the processes utilised and data generated for a voltage-gated cation channel blocker project. The first step in the process was development of a computational pharmacophore model for the ion channel target of interest. In the current example, data pertaining to blockers of voltage-gated cation channels was collated from a number of sources. Historical data was collected from internal AstraZeneca projects, resulting in a knowledge base that was expanded by data mining in patent and literature sources. For example, analysis of the scientific literature revealed 170 small molecules that interact with either voltage-gated sodium, calcium or potassium channels. Data were then filtered according to the experimental method used, with patch-clamp concentration-response data given the highest weighting. Using the final compound list, a 3-dimensional pharmacophore model was generated using computational methods.
In order to select compounds for synthesis, extensive in silico screening was carried out against the 3D pharmacophore model of our voltage-gated cation channel target. The input for this screening was supplied by generating ideas targeting ion channels then enumerating these ideas into ‘virtual’ libraries. To date, we have generated a ‘virtual’ library of more than 107 chemical structures that are amenable to multiple parallel synthesis chemistry. From the results of the in silico screening, compounds were scored according to their fit to the pharmacophore model. The best virtual library was selected and a sub-set of compounds was finally synthesised. Purification of the synthesised compounds results in a focussed library of several hundred compounds that can then be passed on to bio-scientists for testing.
In order to analyse the focussed chemical libraries generated through the above process, we chose to utilise one of the new automated electrophysiology platforms. This approach is particularly important for voltage-gated ion channels where transition between conformational states is regulated by membrane potential and depolarisation of the cell is the physiological stimulus for channel opening. After consideration of the various available technologies, we selected the IonWorks platform, which is a proven technology for screening voltage-gated ion channels4. Two instruments have been marketed by Molecular Devices: IonWorksHT is a single-cell assay based on a 96 well compound plate in combination with a 384 well assay plate. The mismatch between plate formats is important as it provides a level of redundancy such that data is generated for each well of the compound plate. Therefore, in essence IonWorksHT is a 96 well assay and it is possible to test approximately 600 compounds per day at a single-concentration, or 70 compounds per day as 8-point concentration response curves. A second generation instrument, IonWorks Quattro, has recently been launched that makes recordings from up to 64 cells per well and is therefore amenable to screening from a 384 well compound plate. This increases throughput to > 2000 compounds per assay day (at a single concentration), thereby enabling the rapid profiling of a large number of focussed libraries.
Using electrophysiological methods in the primary library-screening phase provides a number of advantages.
- The assay readout – electrical current equivalent to the total flux of ions through all channels in the cell – is a very direct measure of ion channel activity, therefore false positives are rarely encountered. Toxic compounds, which may reduce the functional ion channel response, are detected as a loss in membrane integrity. False negatives are possible due to problems with compound solubility, but in many cases these can be detected through irregularities in the concentration response curve or through visual inspection of the assay plate.
- The patch-clamp technique utilised on IonWorks and other high-throughput electrophysiology platforms, allows control of extra-cellular and intra-cellular solutions and, most importantly, of cellular membrane potential. This means that assay design and data interpretation is not a major problem as it can be when using indirect readouts of ion channel activity, where control of membrane potential is not possible.
- As the electrophysiology readout is in real-time, kinetic information can be collected providing an insight into mechanism of action. For example, many ion channel modulators exhibit the property of ‘state-dependence’, whereby compounds bind preferentially to one or more conformational states. This property is key to the clinical activity of voltage-gated sodium and calcium channel blockers.
- Provided that the same host cell line is utilised, accurate determinations can be made of selectivity between ion channels under very similar assay conditions.
Returning to our voltage-gated cation channel target: as described above, multiple libraries were generated by MPS chemistry. These libraries required biological feedback in order to test the pharmacophore model and to identify hit series to be progressed further. For reasons of throughput, we chose to test library compounds initially at a single-concentration. A screening concentration of 10μM was selected to provide a good compromise between minimising compound usage and reducing solubility issues, while working at a relevant concentration for hit identification. Libraries were screened in parallel against three IonWorks assays; the primary target, a selectivity hurdle and the cardiac hERG potassium channel. The latter channel is associated with drug-induced cardiac arrhythmias5. Therefore, from this initial experiment we were able to build up a biological fingerprint that could be used to prioritise libraries for follow-up analysis. An additional advantage of the screening panel approach is that false positives such as cytotoxic compounds will show similar results in all assays and will therefore not be prioritised for follow-up analysis. The next step was to quantify the activity of hit compounds through concentration-response analysis. As our libraries are generated with relatively conservative variations around a common chemical scaffold, it is possible to generate hypotheses about structure-activity relationships from these data. Finally, we quantified state-dependence for every compound tested, thereby providing an insight into mechanism-of-action. All of this information was then fed back to the computational chemists, who could then refine the pharmacophore models and synthesise new libraries, thereby re-iterating the whole process.
For the voltage-gated cation channel target described above, we have now tested approximately 50 ion channel focussed libraries and have seen activity in more than 25% of the samples tested. From these active compounds, a number of chemical series are currently being progressed forward in the drug discovery process, with several back-up series also available. Interestingly, the hit rate recorded in this focussed screening campaign was 100-fold greater than that seen for the same ion channel target in a full scale, random HTS. We therefore believe that the application of in silico screening followed by focussed library synthesis provides a very effective route to ion channel drug discovery. Furthermore, this approach provides significant savings in both cost and time.
In the example described here, a wealth of background pharmacological information was available on the targeted ion channel sub-family. However, for other ion channel targets this may not be the case and therefore application of virtual screening may not be feasible. One solution to this problem is to run a conventional, random screen and then use the resulting hit compounds to develop a computational pharmacophore model and follow up with a virtual screen. Another exciting possibility arises through the recent advances in ion channel structural determination. Since 1998, X-ray structures have been solved for several bacterial ion channels and earlier this year MacKinnon and co-workers reported the first X-ray crystal structure for a mammalian ion channel, the rat Kv1.26 potassium channel. This breakthrough will facilitate the development of structural homology models for voltage-gated cation channels and one can therefore envisage using a combination of structure-based and pharmacophore-based computational models to drive ion channel drug discovery. In the longer term, further progress in ion channel structural biology may allow co-crystallisation of the target ion channel and small molecule modulator, thereby initiating the era of truly rational ion channel drug design.
Acknowledgements
The authors would like to thank Olof Larsson, Graeme Wilkinson, John Ridden and Karl Hard for their valuable input to this manuscript.

References
- Obituary, Leo Sternbach. The Guardian, 3rd October 2005.
- Wafford, K.A. (2005) GABAA receptor sub-types: any clues to the mechanism of benzodiazepine dependence? Curr.Opin.Pharmacol. 5(1):47-52.
- Trezise D.J. (2005) Patch clamp electrophysiology steps up a gear. Eur. Pharmaceutical Rev. 2:69-73.
- Schroeder K, Neagle B, Trezise D.J. & Worley J (2003). IonWorks HT: a new high-throughput electrophysiology measurement platform. J.Biomol.Screen. 8: 50-64.
- Sanguinetti M.C. & Mitcheson J.S. (2005). Predicting drug-hERG channel interactions that cause acquired long QT syndrome. Trends Pharmacol Sci. 26(3):119-24
- Long S.B., Campbell E.B. & Mackinnon R. (2005) Crystal structure of a mammalian voltage-gated Shaker family K+ channel. Science 309(5736):897-903.




