Developing and applying recombinant antibody microarrays for high-throughput disease proteomics
Posted: 16 December 2010 |
Deciphering crude proteomes in the quest for candidate biomarker signatures for disease diagnostics, prognostics and classifications has proven to be challenging using conventional proteomic technologies. In this context, affinity protein microarrays, and in particular recombinant antibody microarrays, have recently been established as a promising approach within high-throughput (disease) proteomics1-3. The technology will provide miniaturised set-ups capable of profiling numerous protein analytes in a sensitive, selective and multiplexed manner.
The microarray patterns generated can then be transformed into proteomic maps, or detailed molecular fingerprints, revealing the composition of the proteome at a molecular level. By using bioinformatics, biomarker signatures can then be de-convoluted and tested for clinical applicability and in the end hopefully be transferred into clinical practice. In this mini-review, we will discuss selected steps in the process of developing and applying recombinant antibody microarrays for high-throughput disease proteomics, as well as addressing the issue of transferring proteomic discoveries into clinical practice.
Antibody microarrays – definition
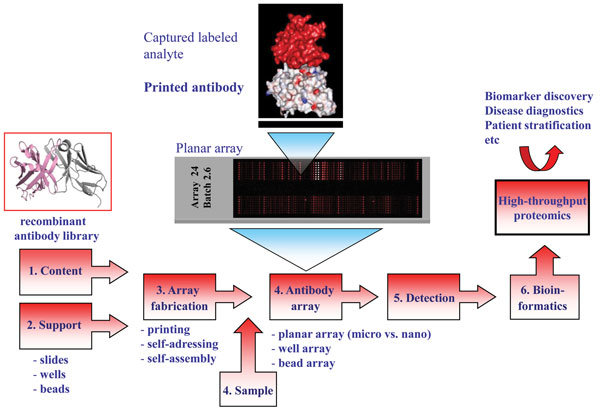
Entering the post-genomic era sparked the development of technology platforms for protein expression profiling, including protein microarrays and in particular antibody-based arrays1,2,4-6. The idea of generating miniaturised micro and nano-sized (mm2 to cm2 range) antibody arrays is based on direct printing (≤ pL scale)1,5,7,8, self-addressing9-11 or selfassembling12- 16 minimal amounts (fmol range) of individual antibodies (ranging from a few to several hundreds) with the desired specificity in discrete positions (nm to μm sized spot features) in an ordered pattern, an array, onto a solid support (Figure 1). While planar affinity arrays on slides constitute the dominating layout, multiplexed arrays have also been produced on the bottom of ELISA plate wells as well as in solution, so-called bead arrays2,17-20. The immobilised antibodies will act as highly specific capture molecules, or probes, for the targeted analytes. The arrays are then exposed to small amounts (μL scale) of (labelled) non-fractionated proteomes, e.g. serum, plasma, urine, tissue extract or cell lysates21, before specifically bound analytes are detected and quantified, mainly using fluorescence as mode of sensing. These multiplexed and rapid (< three hours) analyses regularly display an assay sensitivity in the pM to fM range, enabling also low-abundant (pg/ml) analytes to be targeted in crude proteomes22-24. The array images are then transformed into protein expression profiles, or protein atlases, de-convoluting the composition of the sample at a molecular level. Finally, bioinformatics could be applied to define differences and similarities in the protein expression profiles between the sample cohorts at hand, e.g. healthy vs. cancer, resulting in the discovery of e.g. potentially disease-associated protein analytes, or candidate biomarker signatures.
Why antibody microarrays – from focused assays to immunosignaturing
Taking advantage of the explicit specificity displayed by high-performing antibodies, antibody-based arrays are highly suitable for protein expression profiling efforts, ranging from focused assays targeting a handful of analytes to proteome wide screenings1,2,8. In contrast to traditional proteomic technologies, such as mass spectrometry, protein microarrays have proven capable of targeting crude, non-fractionated proteomes in a selective, specific and sensitive manner representing key technological advantages1,2,8.
The number and range of specificities included on the array will set the stage for the application at hand2. Depending on the application, such as disease diagnostics, this choice will be critical for the end result. One approach could be to explore the wealth of information already available to identify candidate markers, for which antibodies are then generated and included on the array. Another solution would be to first perform a discovery study, based on e.g. gene expression profiling, to identity the candidate markers25. Focusing on biomarker discovery for disease diagnostics and prognostics, we have chosen a different approach to this key step (Figure 2). Using affinity proteomics, we have decided to harness the diagnostic/prognostic power of the immune system to target various indications, focusing on cancer and inflammatory indications26-30 (Wingren et al., unpublished observations). We have based our approach on the well-established notion that the immune system is remarkably sensitive to changes in an individual’s state of health, resulting from disease, registering these alterations through fluctuations in the levels of various analytes, in particularly immunoregulatory analytes. To this end, we have designed a recombinant antibody microarray technology platform targeting mainly immunoregulatory proteins2,22,24. These immunosignatures could be considered as snapshots of the immune system’s activity in a patient at the time of the test, giving us a unique read-out reflecting what is going on inside the patient. These fingerprints will reflect a combination of direct and indirect (systemic) effects in response to the disease, and could collectively be explored and exploited as candidate classifiers for the disease. Albeit the range of specificities could still be a limiting factor (in our case, we have regularly used a few hundred antibodies to target ≤80 analytes), we have still demonstrated that disease-associated candidate biomarker signatures displaying high diagnostic and prognostic power could be deciphered2,26-31, outlining the strength of the technology. In the end, de-convoluting the composition of complexes proteomes, such as serum, in both health and disease, will generate novel biomarker panels which could be utilised within personalised medicine – setting the stage for the next generation of healthcare.
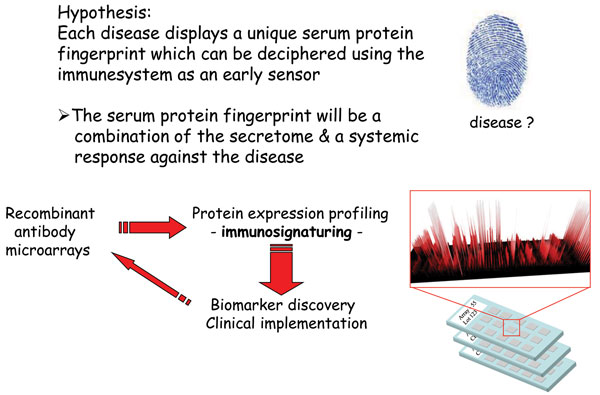

Figure 2 Immunosignaturing – harnessing the diagnostic / prognostic power of the immune system using affinity proteomics
Design and development of antibody microarrays
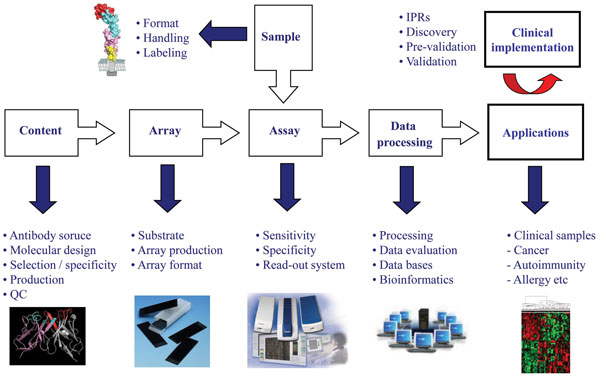
The process of developing high-performing antibody micro and nano-arrays has turned out to be demanding, requiring truly crossdisciplinary approaches (e.g. surface chemistry, protein engineering, bio and nanotechnology, analytical sciences and bioinformatics) to be adopted1,2,4-6,8,17,23. In our own efforts to design recombinant antibody microarrays, early on we identified five key technological issues, including i) content, ii) array design, iii) sample format, iv) assay design, and v) data processing, that had to be addressed in parallel in order to succeed (Figure 3). These key areas and the associated processes have been reviewed elsewhere and will therefore not be addressed here in general terms2,8,22,24,32. Instead, we have selected a few key points that will be highlighted and illustrated, focusing on the content (antibody design and range of specificities) and array design (micro vs. nano).
The design of the antibodies used as probes will be essential as it directly affects and/or reflects the i) on-chip performances, ii) range of specificities, iii) renewability, iv) scaling-up, and v) molecular design. Based on these five key points, it is thus vital to select the antibody design that best matches the specific requirements of the application at hand. While monoclonal and polyclonal antibodies have worked well in array-based applications4,25,33, it will be difficult to meet all of the above criteria using these probe formats1,2. We and others have shown that recombinant antibodies / antibody libraries constitute a highly attractive readily renewable probe source for antibody-based arrays1-3,8. The libraries can be tailored by protein engineering to generate probes that are microarray adapted by molecular design, displaying high on-chip performances. Furthermore, by using large libraries, composed of up to 2 x 1010 clones, numerous antibodies carrying ‘any desired’ specificities could be selected and included on the array in a manner also compatible with large scale endeavours. Using, as in our case, a human recombinant single-chain Fv (scFv) antibody library genetically constructed around a single, constant framework34 further promotes the possibility to customise the antibodies to perform well in array applications as well as enable (facilitate) the possibility to re-design the scaffold for all probes in a facile manner in order to perform even better.
Despite all advances, the traditional approach of generating and including at least one antibody per analyte poses a significant logistical issue when aiming for a large-scale effort1,2. The need for high-density arrays, i.e. numerous of antibodies, when aiming for global proteomics could, however, be circumvented by adopting the Global Proteome Survey (GPS) approach, a new affinity proteomic discovery platform35. GPS will provide us with unique means to ultimately perform global proteome analysis using a limited number of binders35 (Olsson et al. unpublished observations). The concept is based on interfacing affinity proteomics with a mass spectrometry-based read-out. Briefly, we have selected scFv antibodies against a set of four or six amino acid long peptide motifs, each motif being shared among five to 100 different proteins. These context independent motif specific (CIMS) antibodies could then be used to target motif containing peptides in a specie independent manner. In this way, 200 antibodies, each targeting a motif shared among 50 different proteins, would potentially target almost half the non-redundant human protein. From a practical point of view, the target proteome would first be digested, exposed to immobilised CIMS antibodies, whereby motif containing peptides would be enriched. Next, eluted peptides would be detected and identified using tandem mass spectrometry allowing us to backtrack the original protein even in a quantitative manner. In other words, this approach would significantly reduce the number of antibodies required, by using antibodies specific for a set of proteins, instead of one unique probe per protein as in classical affinity proteomic.
Finally, when aiming for large-scale set-ups, the size of conventional microarrays will become a limiting factor paving the way for antibody-based nanoarrays32,36. While proof-of-concept antibody nanoarrays have been generated for nanosized features32,36,37, the current trend is that sub-micron sized features (about five to ten microns) will be more than sufficient. The advantages of using spot features of this size is that i) conventional scanners with e.g. 0.5 micron resolution, can still be used, ii) adequate sensitivity will be maintained, and iii) sufficient array densities (>200,000 spots/cm2) can still be obtained. To this end, nanospotters relying on dip pen technology or microcantilever methodology have been developed, allowing multiplexed arrays to be printed, setting the stage for the next generation of miniaturised array assays consuming minute amounts of reagents and sample.
Antibody microarray-based applications
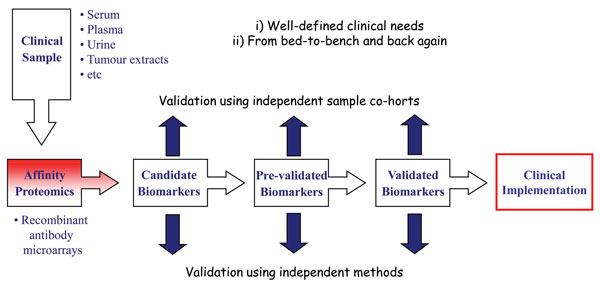
To date, a wide variety of applications have been performed using antibody microarrays and in particular recombinant antibody micro – arrays1,2,4,5,7,8,17,20. Focusing on our in-house designed recombinant scFv antibody arrays, we have demonstrated the applicability of our technology platform within disease proteomics, focusing on cancer (e.g. breast cancer, pancreatic cancer, gastric adenocarcinoma and multiforme glioblastoma) and inflammatory conditions (e.g. systemic lupus erythematosus, systemic sclerosis, pre-eclampsia and Helicobacter pylori infection)22,26-30,38 (Wingren et al., unpublished observations). In these efforts, we have first organised the project around a well-defined clinical need, and based on this, planned the work in a truly translational manner, going from the bed-to bench and back again (Figure 4). In order to adopt a stringent biomarker discovery program, it is important that sequential studies are planned, going from discovery, pre-validation to validation phase, each step involving a new, independent patient data set to be targeted. Further, the findings reported in each phase should also be cross-validated using alternative, parallel methods.
Adopting this work-flow, we have deciphered several candidate biomarker signatures in crude proteomes, such as serum, plasma and tissue extracts. Some of these projects have also entered the (pre-)validation phase, enabling us to report some of the first pre-validated, multiplexed biomarker signatures based on proteomics. Briefly, we have identified candidate biomarker signatures enabling us to discriminate metastatic breast cancer, gastric adenocarcinoma and pancreatic cancer from healthy controls displaying a high diagnostic power27,29,30. In the case of pancreatic cancer, a candidate signature indicating survival was also indicated. No validated blood-test for diagnosis and prognosis of pancreatic cancer, displaying a five year survival of less than five per cent yet exists clearly highlighting the clinical need. Furthermore, we have pinpointed candidate plasma biomarker signatures associated with prognosis and therapy selection in glioblastoma multiforme patients26. Finally, using affinity proteomics, we have taken one step further towards de-convoluting the complex features of pre-eclampsia at the molecular level28. Targeting placenta tissue extracts, we have outlined the first generation of candidate pre-eclampsia associated placenta tissue protein signatures, and indicated that pre-clampsia and subgroups thereof could be distinguished.
Transferring proteomic discovery intro clinical practice
The field of proteomics demonstrates great promise for providing improved means for early disease diagnostics, prognosis, patient stratification, monitoring the effects of therapy and predicting the risk for relapse. To this end, a variety of proteomic methodologies has been developed and implemented in the quest to decipher candidate biomarker panels. For several years, numerous studies have also been published, outlining the discovery of such tentative biomarker panels in cells, tissues and/or body fluids. Despite these exciting reports, the subsequent efforts of transferring these proteomic discoveries into clinical practice, providing validated signatures of clinical relevance have, however, yet to deliver39. The reasons behind this, so far, impaired translation are manifold and complex, including e.g. technical limitations, sample handling, study quality, difficulties in implementing the multidisciplinary approaches required as well as issues of validating earlier findings. These key issues highlight the challenges that clinical proteomics must face before its full potential can be explored and exploited. But considering recent advances, the opportunities for translational research going from bedside to bench and back again is finally looking more promising, and adopting state-of-the-art proteomic tools show improved promise for deciphering clinically relevant candidate biomarker panels that then can be transferred into clinical practice.
Ackowledgements
This study was supported by grants from the Swedish National Research Council (VR-NT and VR-M), the Foundation of Strategic Research (Strategic Center for Translational Cancer Research-CREATE Health (www.createhealth.lth.se), Crafoord foundation, Vinnova, Alfred Österlund foundation and Great and Johan Kocks foundation.
References
- Borrebaeck, C.A.K. & Wingren, C. (2007) High-throughput proteomics using antibody microarrays: an update. Exp Rev Mol Diagnos 7, 673-686
- Borrebaeck, C.A.K. & Wingren, C. (2009) Design of high-density antibody microarrays for disease proteomics: Key technological issues. J Proteomics
- Pavlickova, P., Schneider, E.M. & Hug, H. (2004) Advances in recombinant antibody microarrays. Clin Chim Acta 343, 17-35
- Haab, B.B. (2002) Applications of antibody array platforms. Curr Opin Biotechnol
- MacBeath, G. Protein microarrays and proteomics. Nat Genet 32 Suppl, 526-532 (2002).
- Wingren, C. & Borrebaeck, C.A.K. (2009) Antibody-based microarrays. Methods Mol Biol (Clifton, N.J) 509, 57-84 7. Angenendt, P. (2005) Progress in protein and antibody microarray technology. Drug Discov Today 10, 503-511
- Wingren, C. & Borrebaeck, C. (2006) Antibody Microarrays – Current Status and Key Technological Advances. OMICS 10, 411-427
- Svedhem, S., et al. (2003) Patterns of DNA-labeled and scFv-antibody-carrying lipid vesicles directed by material-specific immobilization of DNA and supported lipid bilayer formation on an Au/SiO2 template. Chembiochem 4, 339-343
- Wacker, R. & Niemeyer, C.M. (2004) DDI-microFIA–A readily configurable microarray-fluorescence immunoassay based on DNA-directed immobilization of proteins. Chembiochem 5, 453-459
- Wacker, R., Schroder, H. & Niemeyer, C.M. (2004) Performance of antibody microarrays fabricated by either DNA-directed immobilization, direct spotting, or streptavidin-biotin attachment: a comparative study. Anal Biochem 330, 281-287
- He, M., et al. (2008) Printing protein arrays from DNA arrays. Nature methods 5, 175-177
- He, M., Stoevesandt, O. & Taussig, M.J. (2008) In situ synthesis of protein arrays. Curr Opin Biotechnol19, 4-9
- Ramachandran, N., et al. (2004) Self-assembling protein microarrays. Science 305, 86-90
- Ramachandran, N., Hainsworth, E., Demirkan, G. & LaBaer, J. (2006) On-chip protein synthesis for making microarrays. Methods Mol Biol (Clifton, N.J) 328, 1-14
- Ramachandran, N., et al. (2008) Next-generation high-density self-assembling functional protein arrays. Nature methods 5, 535-538
- Hartmann, M., Roeraade, J., Stoll, D., Templin, M.F. & Joos, T.O. (2009) Protein microarrays for diagnostic assays. Analytical and bioanalytical chemistry 393, 1407-1416
- Joos, T.O., Stoll, D. & Templin, M.F. (2002) Miniaturised multiplexed immunoassays. Curr Opin Chem Biol 6, 76-80
- Schwenk, J.M., Gry, M., Rimini, R., Uhlen, M. & Nilsson, P. (2008) Antibody suspension bead arrays within serum proteomics. J Proteome Res 7, 3168-3179
- Templin, M.F., Stoll, D., Bachmann, J. & Joos, T.O. (2004) Protein microarrays and multiplexed sandwich immunoassays: what beats the beads? Comb Chem Highthroughput Screen 7, 223-229
- Wingren, C. & Borrebaeck, C.A.K. (2008) Antibody microarray analysis of directly labelled complex proteomes. Curr Opin Biotechnol 19, 55-61
- Ingvarsson, J., et al. (2007) Design of recombinant antibody microarrays for serum protein profiling: targeting of complement proteins. J Proteome Res 6, 3527-3536
- Kusnezow, W., et al. (2007) Antibody microarray-based profiling of complex specimens: systematic evaluation of labeling strategies. Proteomics 7, 1786-1799
- Wingren, C., Ingvarsson, J., Dexlin, L., Szul, D. & Borrebaeck, C.A.K. (2007) Design of recombinant antibody microarrays for complex proteome analysis: choice of sample labeling-tag and solid support. Proteomics 7, 3055-3065
- Sanchez-Carbayo, M., Socci, N.D., Lozano, J.J., Haab, B.B. & Cordon-Cardo, C. (2006) Profiling bladder cancer using targeted antibody arrays. Am J Pathol 168, 93-103
- Carlsson, A., et al. (2010) Plasma proteome profiling reveals biomarker patterns associated with prognosis and therapy selection in glioblastoma multiforme patients. Proteom Clin Appl 4, 591-602
- Carlsson, A., et al. (2008) Serum proteome profiling of metastatic breast cancer using recombinant antibody microarrays. Eur J Cancer 44, 472-480
- Dexlin-Mellby, L., et al. (2010) Tissue protome profiling of preeclamptic placenta tissue using recombinant antibody microarrays. Proteomics – Clin Applic 4, 794-807
- Ellmark, P., et al. (2006) Identification of protein expression signatures associated with Helicobacter pylori infection and gastric adenocarcinoma using recombinant antibody microarrays. Mol Cell Proteomics 5, 1638-1646
- Ingvarsson, J., et al. (2008) Detection of pancreatic cancer using antibody microarraybased serum protein profiling. Proteomics 8, 2211-2219
- Dexlin, L., et al. (2006) Membrane protein profiling of intact cells using recombinant antibody microarrays. J Proteome Res
- Wingren, C. & Borrebaeck, C.A.K. Progress in miniaturization of protein arrays–a step closer to high-density nanoarrays. Drug Discovery Today 12, 813-819 (2007).
- Haab, B.B. (2005) Antibody arrays in cancer research. Mol Cell Proteomics 4, 377-383
- Soderlind, E., et al. (2000) Recombining germline-derived CDR sequences for creating diverse single-framework antibody libraries. Nat Biotechnol 18, 852-856
- Wingren, C., James, P. & Borrebaeck, C.A.K. (2009) Strategy for surveying the proteome using affinity proteomics and mass spectrometry. Proteomics 9, 1511-1517
- Wingren, C., Montelius, L. & Borrebaeck C, A.K. (2003) Protein Microarrays Chapter 17, 339-352
- Ghatnekar-Nilsson, S., Dexlin, L., Wingren, C., Montelius, L. & Borrebaeck, C.A.K. (2007) Design of atto-vial based recombinant antibody arrays combined with a planar wave-guide detection system. Proteomics 7, 540-547
- Strachan, R.T., Ferrara, G. & Roth, B.L. (2006) Screening the receptorome: an efficient approach for drug discovery and target validation. Drug Discovery Today 11, 708-716
- Borrebaeck, C.A.K. & Wingren, C. (2009) Transferring proteomic discoveries into clinical practice. Exp Rev Proteomics 6, 11-13
About the Author
Dr. Wingren is a lecturer at the Department of Immunotechnology, Lund University, Sweden. Currently, Dr. Wingren directs a research group working on the development of recombinant antibody microarrays for high-throughput (disease) proteomics, with a particular focus on oncoproteomics. In 2003, he was promoted to Associate Professor in Immunotechnology. Prior to joining Immunotechnology in 1999, Dr. Wingren pursued his postdoctoral training in structural biology in the laboratory of Prof. Ian Wilson at the Scripps Research Institute, La Jolla, USA (1997-1999). Dr. Wingren received his Ph.D. in biochemistry at Lund University in 1997, and a B.Sc. in chemistry in 1991. Dr. Wingren has published over 75 publications and reviews in peer-reviewed international journal and is co-author on several patent applications.
Contact the Author email: [email protected]
Issue
Related topics
HCS (High Content Screening), HTS (High Throughput Screening), Microarrays, Proteomics