The determination of structural changes of biopharmaceuticals during Freeze-Drying using Fourier Transform Infrared Spectroscopyb
Posted: 20 March 2009 | Heiko A. Schiffter (University of Oxford), Sebastian Vonhoff (University of Erlangen-Nuremberg) | No comments yet
Peptides and proteins are powerful active therapeutic ingredients used in a wide variety of serious conditions and illnesses such as diabetes, arthritis or cancer. The application of these so-called biopharmaceuticals has been rapidly increasing since the middle of the 1990s, facilitated by improvements in modern recombinant DNA technology and biotechnological manufacturing. The worldwide sales of the biotech drug market grew from 43 billion US$ in 2003 to over 75 billion US$ in 2007 according to a recent IMS Health market analysis. The major challenge in the development of stable protein formulations and dosage forms is to ensure their process and shelf life stability.
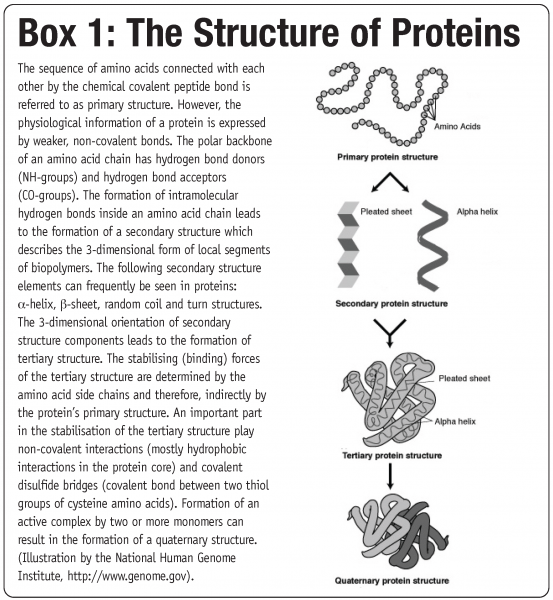
The fact that these biological macromolecules have several levels of structure, referred to as primary, secondary, tertiary and quaternary structure (see Box 1), constitute the basis for multiple interaction pathways that can lead to protein instability1. Damage to at least one type of these substructures is referred to as denaturation and can greatly reduce protein functionality2. As water supports many degradation pathways (e.g. hydrolysis, oxidation or deamidation), long term storage stability of aqueous protein solutions is often limited3. The primary approach to elongate their shelf life is to reduce water availability and activity. This can be achieved to some extend by freezing, but the most desirable approach is to completely remove water and transfer the aqueous protein formulation into a dry powder form.

Lyophilization and Stability of Proteins
Many proteins are sensitive to heat and elevated temperature, and therefore freeze-drying has been the process of choice to accomplish acceptable shelf life for protein formulations and to ensure high quality standards. Freeze-drying or “lyophilization” is a drying process where the solvent, usually water, is first frozen and then removed by sublimation at low temperature and pressure. However, freeze-drying itself can have a negative influence on the structural stability of proteins4. A large number of hydrogen bonds, hydrophobic interactions and Van der Waal forces stabilise the secondary, tertiary and quaternary structure of a protein. Any addition of energy to or the removal of water from this system can cause a disruption of the stabilising forces and allow the protein to unfold, denature and aggregate. During freeze-drying this can happen as a result of (i) cold denaturation due to low temperatures, (ii) , freeze-concentration and related effects such as solute precipitation, pH-shifts or phase separation, (iii) adsorption of proteins to ice/water interfaces due to their hydrophobic/hydrophilic character, and (iv) the removal of water5-9. It is discussed that proteins in aqueous solution have a monolayer of water covering their surface, the so-called hydration shell10. The amount of water in full hydration is 0.30-0.35 g per gram protein11. During lyophilization, most of the water is already removed from the protein during freezing of the aqueous formulation and therefore, most of the drying is actually performed during the freezing process12. Freezing and subsequent ice sublimation can remove parts of the hydration shell. In a water-poor environment, a protein tends to transfer protons to ionized carboxyl groups to abolish the charges in the protein. A decrease in charge density can result in an increase in protein-protein hydrophobic interactions after unfolding and thus, protein aggregation10. If water molecules are an integral part of the active site of a protein with enzymatic activity the removal of these functional water molecules can easily have an impact on residual enzyme activity13. Stable formulation and process design is required to avoid biological activity loss and possible immunogenic effect of the structurally and/or chemically altered proteins molecules. Addition of cryoprotectants and lyoprotectants greatly reduces protein damage and helps preserving its structure and thereby its activity14. To formulate the macromolecular therapeutics, excipients are added into the solution systems to protect the functionality of the proteins and to improve the handling of the final products. Among the common excipients, polyols (e.g. saccharides and sugar alcohols) are effective protectants to many proteins4. Polyols can protect the native conformation of a protein against aggregation and precipitation during freeze-concentration as well as dehydration stresses by replacing essential water molecules through molecular interactions (hydrogen bonding) with the biomolecules4. Additionally, some of these polyols improve the stability of the proteins during the freeze-drying process and subsequent storage by embedding them into a highly viscous glass-state amorphous solid with limited molecular mobility12. Other expicients including polymers, surfactants or inorganic salts can also be important to a successful formulation.
Effects like unfolding and aggregation during freeze-drying are known to directly correlate with protein secondary and tertiary structure15. Methods to examine changes in secondary structure include infra-red spectroscopy or circular dichroism. For the determination of changes in tertiary structure, fluorescence spectroscopy is often applied. It has been found that the degree of retention of native secondary structure in the dried state is an important criterion for the prediction of long term storage stability of proteins in freeze-dried formulations16-18. If spectral changes were only due to removal of water, then every spectrum would exhibit similar changes during lyophilization which is not the case19. The determination and quantification of structural changes during lyophilization can be used to identify the damage caused to biopharmaceuticals during freeze-drying. By linking formulation properties and process conditions to protein native structure, it is possible to derive guidance leading to optimal formulation and process protocol.
Fourier Transform Infrared Spectroscopy of proteins
Identification and characterisation of changes in secondary structure during lyophilization are technically challenging, in particular due to the desire to examine the liquid formulation prior to freeze-drying as well as the dry and rehydrated final product. Fourier Transform Infrared Spectroscopy (FTIR) is the method of choice to obtain qualitative and quantitative information about the secondary structure elements of proteins in different states of aggregation18. For the examination of protein pharmaceuticals in freeze-drying formulation and process development, there are basically three different standard setups for sample spectrum recording:
Transmission cells offer a good signal-to-noise ratio and the most native-like environment for protein samples. They can only be used for liquid samples allowing comparisons between the untreated, freeze-thawed and the rehydrated formulation. Due to the strong IR-absorption of water, measurements must be taken at very low optical pathlengths (<10 μm required, typically 6-8 μm). Alternatively, water can be replaced by D2O.
With attenuated total reflectance (ATR) the IR-beam is reflected multiple times within a crystal carrying the sample. Absorption occurs each time the beam reaches the crystal-sample-interface and optical pathlength is defined by the number of reflections within the crystal. ATR-technology is suitable for recording liquid and solid samples. However, as proteins tend to accumulate at interfaces, spectra of liquid protein samples can exhibit changes in secondary structure due to adsorption to the ATR crystal20.
Pellets/discs were often used to record solid samples before the breakthrough of ATR technology. The protein sample is mixed with KBr and subsequently subjected to high pressure. There are contrary reports about the impact of this procedure on protein stability16,19. Nevertheless, this method is time consuming and not often used anymore these days.
Spectra taken from liquid formulations prior to freeze-drying can be used to evaluate the stability of proteins in solution by testing the influence of pH, ion strength or excipients as well as performing accelerated stability tests at various temperatures. New developments of ATR sample cells, such as the BioATR II© from Bruker Optics, enable the observation of denaturation kinetics and the melting point of one protein in different formulations by applying controlled fixed temperature ramps. Analysis of the lyophilized dry and/or rehydrated product with the initial liquid solution (prior to freeze-drying) can be used to determine structural changes caused by dehydration stress15,21. Even an eventually occurring refolding of the protein after rehydration of a freeze-dried product is detectable22.
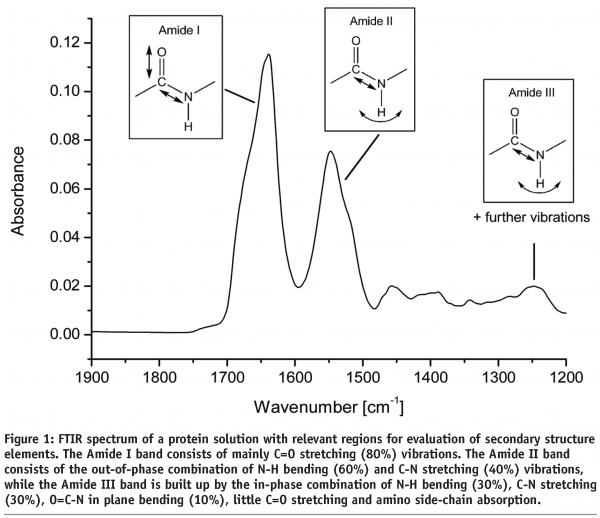

There are three relevant areas within the FTIR spectrum that contain information about protein secondary structure (see Figure 1). The Amide I band is most often used for protein secondary structure examinations as there is plenty of information available in literature and only few molecule vibrations (C=O stretching and little C-N stretching vibrations) contribute to the band thereby facilitating spectral interpretation. On the downside, water absorption is very strong in this region which can lead to noisy and poor reproducible spectra. The Amide II band (arising from N-H bending and C-N stretching) is mostly used to determine the speed of hydrogen-deuterium exchange and thereby to evaluate the accessibility of the protein. The Amide III band has also been used to determine protein secondary structure. In this particular area (1220 – 1330 cm-1) absorbance from water displays no problem but the weak intensity and complex composition of the Amide III band (C-N stretching, N-H bending, O=C-N in plane bending and absorption of protein side-chains) make interpretations difficult23. Any qualitative and quantitative analysis in this review is focused on the Amide I region of the spectrum.
Ideal recording conditions are of paramount importance. Water vapour inside the spectrophotometer increases spectral noise dramatically and changes in temperature have influence on spectral absorbance. These problems can be avoided by constantly purging the equipment with dry air or gaseous N2 as well as by using temperature controlled accessories that prevent changes of the signal of liquid water in the sample or buffer solution. The equipment should be specifically designed for protein samples to avoid problems like fluctuations in pathlength or cell leakage. Also recording parameters like resolution, mirror speeds, number of spectra, etc. should be carefully adjusted to achieve good signal-to-noise ratio. Finally, background subtraction from raw spectra is a rather subjective operation and can cause artifacts. As most proteins do not show absorbance from 1900 – 1720 cm-1, the most pragmatic approach should be choosing a subtraction factor that results in a flat baseline in this region24. If specific equipment for protein analysis is used, sample concentrations down to 0.1-0.5 mg/ml are possible.
Methods for FTIR spectra evaluation
Depending on the applied evaluation method, the results obtained by FTIR spectroscopy can be highly subjective. As one can see in the following paragraphs, different approaches have been undertaken to quantify secondary structures and their changes, each one of them offering advantages and disadvantages. Correct information can only be obtained by choosing an evaluation method suited for the task at hand with objective parameters.
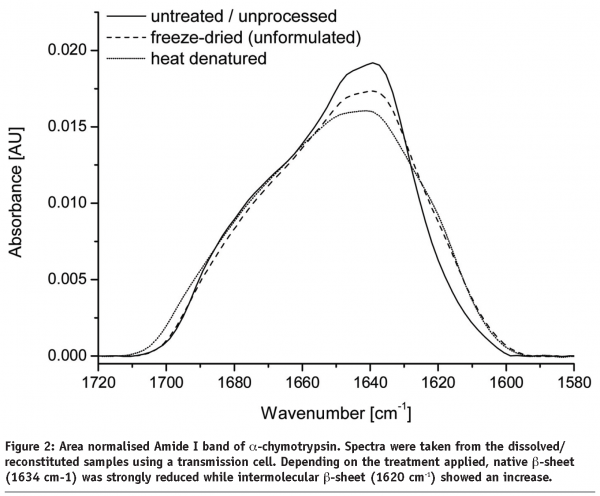

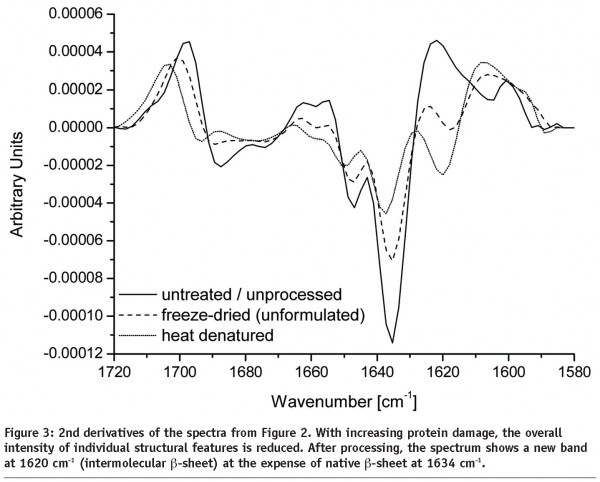

Any changes in protein secondary structure due to lyophilization stresses can be detected by comparing spectra of the untreated and the freeze-dried samples. A straight forward procedure is calculation of the correlation coefficient “r” (see Equation) of the area normalised raw spectra or their 2nd derivatives (see Figures 2 and 3)25,26. The closer the calculated value is to one the better is the match of the two compared spectra. Shifting and broadening of the Amide I band due to lyophilization induced unfolding can easily and objectively be detected by this method6. However, detailed information about changes in protein secondary structure cannot be obtained. Also, a macroscopic comparison of the raw spectra is still necessary to identify false negative results due to baseline effects.
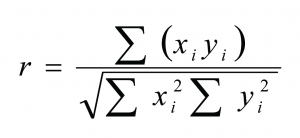
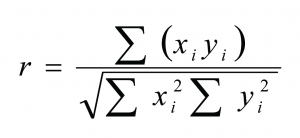
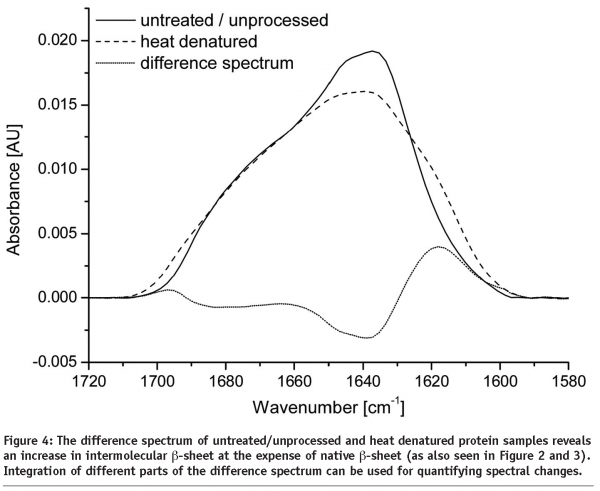
More information concerning structural changes can be obtained by subtracting the baseline corrected, area normalised Amide I bands of the protein spectra (see Figure 4). Any increase or decrease of the Amide I band is visible within the difference spectrum. With knowledge in assigning wavenumbers to secondary structure elements, changes can easily be quantified by putting the integrated areas of the difference spectrum in relation to the area of the raw spectrum. However, information gained remains rather vague because overlapping areas complicate differentiation between individual structural components.

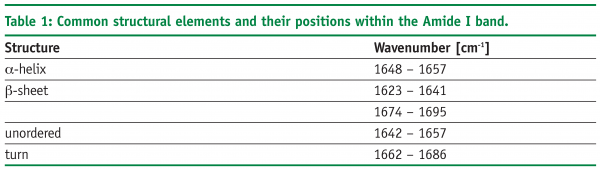

A further approach of detecting structural changes is peak fitting27. Different secondary structure elements show up at defined wavenumbers and their signal correlates with their percentile content within the protein molecule (see Table 1). As the Amide I band consists of multiple superimposing peaks, its shape is rather weak and difficult to interpret. That’s why the spectrum has to be preprocessed before quantification. Fourier Self-Deconvolution (FSD) and 2nd derivative spectra are often used to visualise overlapping peaks within the Amide I region. FSD is a band-narrowing technique that improves the outline of the band and thereby facilitates spectral interpretation (see Figure 5). According to literature, the integrated areas of the component bands remain unaltered and peak fitting can more easily be performed28. However, great care must be taken during FSD as the results are strongly dependent on the factors chosen (bandwidth “w” and exponential “n”) and a high amount of subjectivity is introduced. 2nd derivative spectra, on the other hand, offer an easy and objective way to qualitatively determine peak positions and numbers. Height and width of the individual peaks of the non-deconvolved spectrum can be calculated by an iteration process. Software tries to rebuild the Amide I band based on the assumption that the sum of all peak areas gives the recorded spectrum (see Figure 6). Obviously, this technique is very time consuming and has some disadvantages:
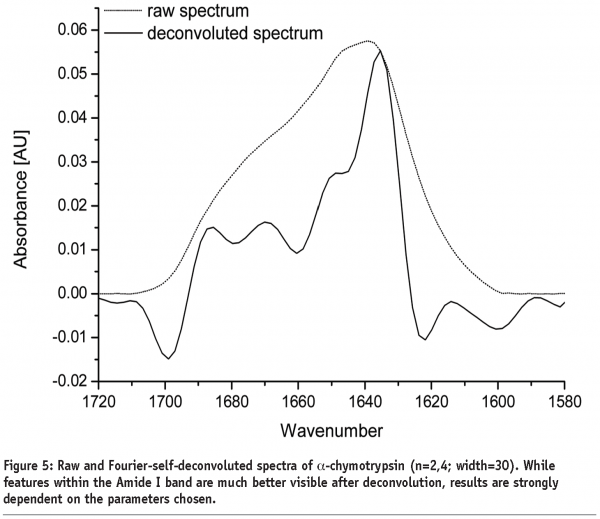

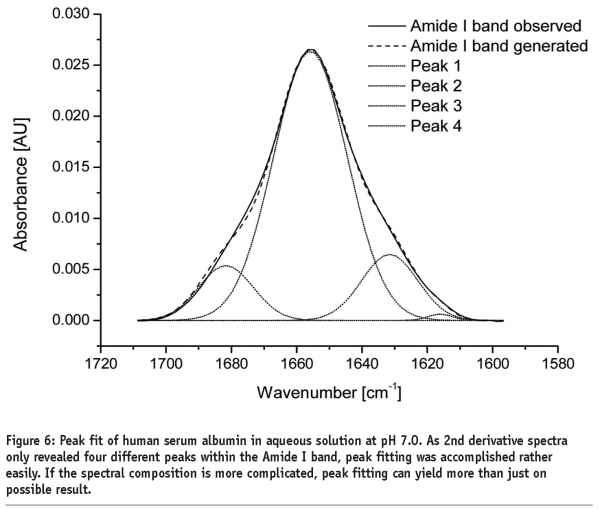

Due to the weak shape of the Amide I band, more than just one peak fit is often ossible
The fit is bound to be incorrect if not all peaks are placed correctly
Wrong assignment of peak positions and corresponding secondary structure elements will lead to false results.
The fourth approach for protein quantification using FTIR spectroscopy is pattern recognition techniques29,30. The sample spectrum is compared with a set of standards with known secondary structure using principal component analysis, singular value decomposition or factor analysis. Failures due to false peak assignment are avoided and structural changes can be quantified objectively. The information obtained is limited by the parameters included in the standards and any failures during calibration will be visible later.
Further developments in FTIR spectroscopy
An interesting development in infrared spectroscopy is the combination of FTIR microscopy techniques with focal plane array (FPA) detector technology (see Figure 7). In contrast to single element detectors, this technology alters the time frame for sample mapping by several orders of magnitude and permits the simultaneously measurements of 4096 spectra in seconds (using a 64 x 64 FPA detector). All 4096 spectra can then be spectroscopically evaluated, which means that the analysis can range from single-band intensity plots to mathematical approaches such as cluster analysis. The information generated will then be reassembled into the original image format, providing distribution plots of the active ingredient and its formulation excipients (see Figure 8). In this manner, the chemical and structural information is directly translated back into the image31,32.


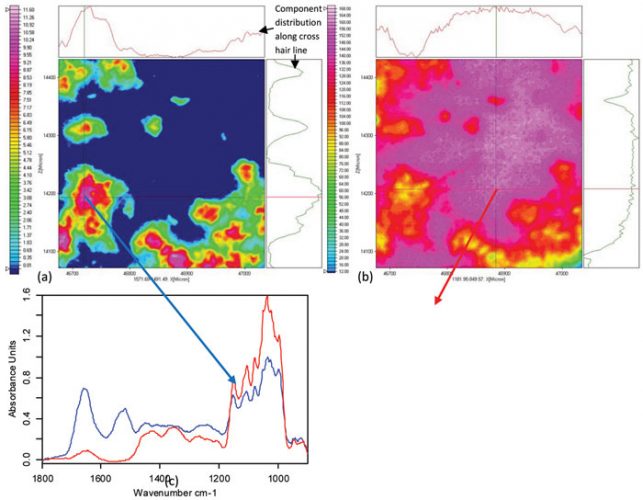

Figure 8: FTIR imaging of lyophilized insulin formulated with trehalose and dextran (100kDa). (a) Image showing the distribution of the protein. Used integration range: 1572 – 1491 cm-1. (b) Image showing the distribution of trehalose/dextran. Used integration range: 1182 – 950 cm-1. (c) Pixel spectra from position of cross-hair: blue shows the amide I and amide II bands of the protein; red shows a spectrum of the formulation components free from protein signals. The lyophilisate was prepared in a diamond cell and measured in transmission. The imaging measurement with a lateral resolution of 2.7µm took three minutes. Imaging measurement and data analysis were performed on a Hyperion 3000 system in collaboration with Bruker Optics, Ettlingen (Germany)
Conclusion
It’s important to realise that FTIR spectroscopy can best be applied for detecting changes in secondary structure. Determination of absolute values by FTIR has always been a challenging task and every quantification method still has its advantages and disadvantages. As long as FTIR spectroscopy is used to assure that no changes occurred during cooling, freezing, freeze-drying or reconstitution, it’s an easy and straight forward procedure. If more information is needed protein FTIR can quickly become more challenging.
Acknowledgements
The authors would like to thank the John Fell Found of the University of Oxford (UK) and Bruker Optics, Ettlingen (Germany) for their support.
References
- Stryer, L.; Berg, J.M.; Tymoczko J.L.; Editors; Biochemistry (2006) 1120 pp.; ISBN 978-0-716-78724-2
- Cox, Michael M.; Phillips, George N., Jr.; Editors; Handbook of Proteins: Structure, Function and Methods; 2 Volume Set (2008) 1378 pp. ISBN 978-0-470-06098-8
- Avis, K.E.; Wu, V.L.; Editors; Biotechnology and Biopharmaceutical Manufacturing, Processing, and Preservation (1996) 400 pp.; ISBN 978-1-574-91016-2
- Wang, W. (2000). Lyophilization and development of solid protein pharmaceuticals. International Journal of Pharmaceutics 203: 1-60.
- Caldarelli, P., De Los Rios, P. (2001). Cold and warm denaturation of proteins. J Biol Phys Vol. 27, No. 2-3, pp. 229-241.
- Pikal-Cleland, K. A. and J. F. Carpenter (2001). “Lyophilization-induced protein denaturation in phosphate buffer systems: monomeric and tetrameric beta- galactosidase.” J Pharm Sci 90(9): 1255-68.
- Bhatnagar, B.S., Pikal, M.J., Bogner, R.H. (2007). Study of the individual contributions of ice formation and freeze-concentration on isothermal stability of lactate dehydrogenase during freezing. J Pharm Sci, Vol. 97, No. 2, pp. 798-813.
- Bhatnagar, B.S., Bogner, R.H., Pikal, M.J. (2007). Protein stability during freezing: Separation of stresses and mechanisms of proteins stabilization. Pharmaceutical Development and Technology 12: 505-523.
- Tang, X. and M. J. Pikal (2004). “Design of freeze-drying processes for pharmaceuticals: practical advice.” Pharm Res 21(2): 191-200.
- Rupley, J. A., Careri, G., 1991. Protein hydration and function. Adv Protein Chem 41: 37-173.
- Kuhlman, B., Yang, H. Y., Boice, J. A., Fairman, R., Raleigh, D. P. (1997). An exceptionally stable helix from the ribosomal protein L9: implications for protein folding and stability. J Mol Biol 270: 640-647.
- Pikal, M.J. (2004) in Freeze-Drying/Lyophilization of Pharmaceutical and Biological Products (Editors: Rey, L. and May, J.C.), pp. 63-107.
- Nagendra, H. G., Sukumar, N., Vijayan, M. (1998). Role of water in plasticity, stability, and action of proteins: the crystal sctructures of lysozyme at very low levels of hydration. Proteins: Struct. Funct. Gen. 32: 229-240.
- Dong, A., Prestrelski, S. J., Allison, S.D., Carpenter, J.F. (1995). Infrared spectroscopic studies of lyophilization- and temperature-induced protein aggregation. J Pharm Sci 84 (4): 415-24.
- Carpenter, J. F., S. J. Prestrelski, Dong, A. (1998). Application of infrared spectroscopy to development of stable lyophilized protein formulations. Eur J Pharm Biopharm 45(3): 231-8.
- Schwegman, J.J., Carpenter, J.F., Nail. S.L. (2007). Infrared microscopy for in situ measurement of protein secondary structure during freezing and freeze-drying. J. Pharm. Sci. 96 (1), pp. 179-195.
- Chang, L., Shepherd, D., Sun, J., Ouellette, D., Grant, K.L., Tang, X., Pikal, M.J. (2005). Mechanism of Protein Stabilization by Sugars During Freeze-Drying and Storage: Native Structure Preservation, Specific Interaction, and/or Immobilization in a Glassy Matrix? J. Pharm. Sci. 94 (7), pp. 1445-1455.
- Carpenter, J.F. (2004), in Freeze-Drying/Lyophilization of Pharmaceutical and Biological Products (Editors: Rey, L. and May, J.C.), pp. 147-186.
- Prestrelski, S.J., Tedeschi, N., Arakawa, T., Carpenter, J.F. (1993). Dehydration- induced conformational transitions in proteins and their inhibition by stabilizers. Biophys J 65 (2), pp. 661-71.
- Fink, D. J., R. M. Gendreau (1984). Quantitative surface studies of protein adsorption by infrared spectroscopy. I. Correction for bulk concentrations. Anal Biochem 139(1): 140-8
- Abdul-Fattah, A. M., D. Lechuga-Ballesteros, et al. (2008). “The impact of drying method and formulation on the physical properties and stability of methionyl human growth hormone in the amorphous solid state.” J Pharm Sci 97(1): 163-84.
- Costantino, H. R., K. Griebenow, et al. (1995). “Fourier-transform infrared spectroscopic investigation of protein stability in the lyophilized form.” Biochim Biophys Acta 1253(1): 69-74.
- Anderle, G. and R. Mendelsohn (1987). “Thermal denaturation of globular proteins. Fourier transform-infrared studies of the amide III spectral region.” Biophys J 52 (1), pp. 69-74.
- Chittur, K.K. (1998). FTIR/ATR for protein adsorption to biomaterial surfaces. Biomaterials 19(4-5): 357-69.
- Kendrick, B.S., Dong, A., Allison, S.D., Manning, M.C., Carpenter, J.F. (1996). Quantification of the area of overlap between second-derivative amide I infrared spectra to determine the structural similarity of a protein in different states. J Pharm Sci 85(2): 155-8.
- Arrondo, J.L., Muga, A., Castresana, J., Goni, F.M. (1993). Quantitative studies of the structure of proteins in solution by Fourier-transform infrared spectroscopy. Prog Biophys Mol Biol 59(1): 23-56.
- Jackson, M., Mantsch, H.H. (1995). The use and misuse of FTIR spectroscopy in the determination of protein structure.” Crit Rev Biochem Mol Biol 30(2): 95-120.
- Susi, H., Byler, D.M., Purcell, J.M. (1985). Estimation of beta-structure content of proteins by means of deconvolved FTIR spectra. J Biochem Biophys Methods 11(4- 5): 235-40
- Jiskoot, W., Crommelin, D.J.A. (2005). Methods for Structural Analysis of Protein Pharmaceuticals. New York, AAPS Press.
- Sarver, R.W., Krueger, W.C. (1991). Protein secondary structure from Fourier transform infrared spectroscopy: a data base analysis. Anal Biochem 194(1): 89-100.
- Schultz, C.P. (2001). Precision infrared spectroscopic imaging. Spectroscopy 16(10): 24-28
- Garidel, P., Boese, M. (2007). Mid infrared microspectroscopic mapping and imaging: a bio-analytical tool for stability and chemically resolved tissue characterization and evaluation of drug permeation within tissue. Microscopy Res and Tech 70, pp. 336-349.



