microRNA manipulation as a host-targeted antiviral therapeutic strategy
Posted: 13 December 2011 |
microRNAs (miRNA) are a class of non-coding RNA that regulate the precise amounts of proteins expressed in a cell at a given time. These molecules were discovered in worms in 1993 and only known to exist in humans in the last decade. Despite the youth of the miRNA field, miRNA misexpression is known to occur in a range of human disease conditions and drugs based on modulating miRNA expression are now in development for treatment of cancer, cardiovascular, metabolic and inflammatory diseases. In the last six years, an increasing number of reports have also illuminated diverse roles of cellular miRNAs in viral infection and a miRNA-targeting therapy is currently in phase II clinical trials for treatment of the Hepatitis C virus. Here we review the literature related to miRNAs that regulate viral replication and highlight the factors that will influence the use of miRNA manipulation as a broader antiviral therapeutic strategy.
microRNAs (miRNA) are a class of small noncoding RNA that bind to messenger RNAs (mRNA) and regulate the amount of specific proteins that get expressed. These small RNAs are derived from longer primary transcripts that fold back on themselves to produce stem-loop structures which are recognised and processed by Drosha and co-factors in the nucleus followed by Dicer and co-factors in the cytoplasm, resulting in a ~ 22 nucleotide (nt) duplex RNA, for review see1,2. One strand of the duplex is preferentially incorporated into the RNA-induced silencing complex (RISC) where it then mediates binding to target mRNAs. These interactions lead to decreased protein getting produced from the transcript, due to RNA destabilisation and/or inhibited translation3 (Figure 1). miRNA-mRNA recognition generally requires perfect complementarity with only the first 6-8 nt of a miRNA, termed the ‘seed’ site4. Each miRNA therefore has the potential to interact with hundreds of target mRNAs3,4 and the majority of human protein-coding genes contain miRNA binding sites under selective pressure5. Therapeutic interest in miRNAs has been supported by studies in model organisms demonstrating key functions of individual miRNAs in cancer, cardiac disease, metabolic disease, neuronal and immune cell function6.
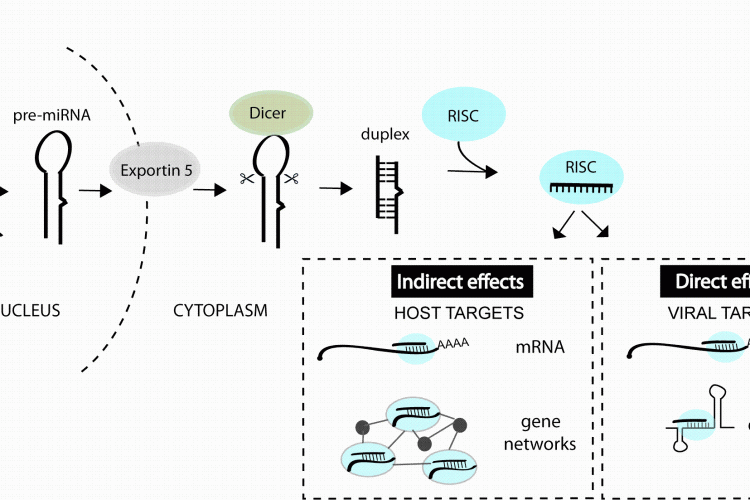

FIGURE 1miRNAs can impact viral infection directly by interacting with viral genes or indirectly by regulating host genes that play a role in the infection. miRNAs are derived from transcripts that contain stem-loop structures which get recognised and processed by a series of enzymes to generate the short (~22 nt) duplex RNA. One strand of the duplex is preferentially incorporated into the RNA-induced silencing complex (RISC) and guides this complex to mRNAs or other viral elements that contain regions of complementarity to the miRNA
microRNAs (miRNA) are a class of non-coding RNA that regulate the precise amounts of proteins expressed in a cell at a given time. These molecules were discovered in worms in 1993 and only known to exist in humans in the last decade. Despite the youth of the miRNA field, miRNA misexpression is known to occur in a range of human disease conditions and drugs based on modulating miRNA expression are now in development for treatment of cancer, cardiovascular, metabolic and inflammatory diseases. In the last six years, an increasing number of reports have also illuminated diverse roles of cellular miRNAs in viral infection and a miRNA-targeting therapy is currently in phase II clinical trials for treatment of the Hepatitis C virus. Here we review the literature related to miRNAs that regulate viral replication and highlight the factors that will influence the use of miRNA manipulation as a broader antiviral therapeutic strategy.
microRNAs (miRNA) are a class of small noncoding RNA that bind to messenger RNAs (mRNA) and regulate the amount of specific proteins that get expressed. These small RNAs are derived from longer primary transcripts that fold back on themselves to produce stem-loop structures which are recognised and processed by Drosha and co-factors in the nucleus followed by Dicer and co-factors in the cytoplasm, resulting in a ~ 22 nucleotide (nt) duplex RNA, for review see1,2. One strand of the duplex is preferentially incorporated into the RNA-induced silencing complex (RISC) where it then mediates binding to target mRNAs. These interactions lead to decreased protein getting produced from the transcript, due to RNA destabilisation and/or inhibited translation3 (Figure 1). miRNA-mRNA recognition generally requires perfect complementarity with only the first 6-8 nt of a miRNA, termed the ‘seed’ site4. Each miRNA therefore has the potential to interact with hundreds of target mRNAs3,4 and the majority of human protein-coding genes contain miRNA binding sites under selective pressure5. Therapeutic interest in miRNAs has been supported by studies in model organisms demonstrating key functions of individual miRNAs in cancer, cardiac disease, metabolic disease, neuronal and immune cell function6.
Diverse functions of miRNAs in viral infection have emerged in the last six years. In 2005, Jopling et al made the seminal discovery that the 5’ end of Hepatitis C virus (HCV) interacts with the liver-specific miRNA, miR-1227. Using a stable cell line that expresses the HCV replicon they showed that inhibition of miR-122 led to 80 per cent reduction in the level of viral replicon RNA, which was later found to similarly reduce the amount of infectious virus8. Although the mechanism for miR-122-dependent enhanced replication is still not clear, just five years after this discovery a miR-122 inhibitor was shown to be successful at reducing viremia in vivo when administered to chimpanzees chronically infected with HCV9 and a miR-122 inhibitor is now in phase II clinical trials10. The pace of progress with the miR-122 inhibitor highlights one of the advantages of miRNAs as drug targets: many are perfectly conserved in animals, enabling the development and testing of miRNA-targeting therapies in various models.
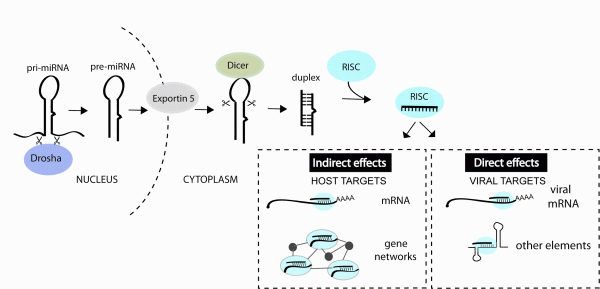

FIGURE 1miRNAs can impact viral infection directly by interacting with viral genes or indirectly by regulating host genes that play a role in the infection. miRNAs are derived from transcripts that contain stem-loop structures which get recognised and processed by a series of enzymes to generate the short (~22 nt) duplex RNA. One strand of the duplex is preferentially incorporated into the RNA-induced silencing complex (RISC) and guides this complex to mRNAs or other viral elements that contain regions of complementarity to the miRNA
Hundreds of potential miRNA binding sites have now been predicted in other viral genomes11-13 but most have not yet been validated (in part owing to the youth of the field). An accumulation of datasets also reveal changes in miRNA expression during infections14 but the functional meaning of most changes also awaits further study. What we do know from the published functional studies is that cellular miRNAs can play very diverse roles in a viral infection, by directly interacting with viral genes or by regulating host genes and pathways involved in the infection process (Figure 1). The purpose of this review is to classify the types of miRNA-virus interactions that occur in mammalian systems and highlight the parameters that influence the antiviral therapeutic capacity of miRNA manipulation.
Direct regulation of viral genes by cellular miRNAs
RNA interference (RNAi) occurs throughout eukaryotic life as a mechanism for the sequencespecific regulation of gene expression15. The unifying feature of RNAi processes is the use of small guide RNAs to direct large RNA-protein complexes to other nucleic acids in order to regulate the expression of genes. Diversity in RNAi-based mechanisms is based on the origin of the guide RNAs, the composition of the RNAprotein complexes and the class of nucleic acids targeted16. RNAi has historically been considered as an antiviral defence mechanism based on studies in insects and plants17. In these systems, the RNAi machinery recognises viral dsRNA and processes it into small RNAs that then guide the nucleases to the viral genome for degradation. Although this antiviral mechanism has not been found in vertebrate systems, in 2005 Lecellier et al. showed that inhibition of the RNAi machinery (which is required for the function of miRNAs) led to enhanced accumulation of Primate Foamy virus-1 (PFV-1) RNA in human cells18. They went on to identify binding sites for several cellular miRNA in the PFV-1 genome and demonstrated that one specific interaction between miR-32 and the PFV-1 3’UTR resulted in a threefold reduction in PFV-1 mRNA. Similar miRNA-virus interactions have now been identified in a human retrovirus: five miRNAs were found to suppress Human Immunodeficiency virus 1 (HIV-1) production in CD4+ T cells19. An additional miRNA, miR-29a, has also been shown to interact with the HIV-1 3’UTR to suppress viral replication20,21 and further miRNAvirus interactions have been reported in human viruses from the Flaviviridae, Hepadnaviridae, Orthomyxoviridae and Rhabdoviridae families (Table 1). In contrast to the miR-122- HCV interaction, all of these other interactions have the expected suppressive effect on viral gene expression, generally resulting in reduced viral replication. In some cases, these antiviral miRNAs are also up- or down-regulated during the infection: for example, miRNAs induced by IFNβ bind to HCV and suppress its replication22. Therapeutic strategies for using these miRNAs would therefore require over-expression or supplementation of the cellular miRNA. However, the evolutionary implications of such antiviral miRNAs merit consideration: if miRNA binding sites were disadvantageous to viruses, a single nucleotide mutation could eliminate this3, so why do viruses maintain binding sites for antiviral cellular miRNAs? One theory is that the down-regulation of viral genes by cellular miRNAs is a mechanism for viral persistence23, consistent with the fact that the miRNAs that suppress HIV are expressed at a high level in CD4+ T cells, which support the latent infection19. Nonetheless, there might still be relevance to using this type of antiviral miRNA in a therapeutic context in a specific stage of the infection, but the potential for resistance is high. On the other hand, another strategy for using miRNAs in a therapeutic context is to harness their natural functions in regulating host genes that viruses require (Figure 1).
TABLE 1 Direct interactions between miRNAs and viruses
| Viral family / Human pathogen | Cellular microRNAs | Function | Reference |
| Flaviviridae | |||
| HCV | miR-122 | Proviral | 7 |
| HCV | miR-196, miR-199a-3p, miR-296, miR-351, miR-431, miR-448 | Antiviral | 22, 44 |
| Hepadnaviridae | |||
| HBV | miR-199a-3p, miR-210, mir-125-5p | Antiviral | 45, 46 |
| Orthomyxoviridae | |||
| Influenza (H1N1) | miR-323, miR-491, miR-654 | Antiviral | 47 |
| Retroviridae | |||
| HIV-1 | miR-150, miR-125-5p, miR-28, miR-223, miR-382, miR-32, miR-29a | Antiviral
| 19-21 |
| Rhabdoviridae | |||
| VSV | miR-24, miR-93 | Antiviral | 48 |
miRNA-modulation of host genes involved in infection
Viruses are parasites whose life cycles are intricately tied to host cell processes. Since miRNAs regulate numerous aspects of host cell physiology (e.g. cell cycle, metabolism, differentiation), their expression levels also impact the infection process. One of the first demonstrations of the relevance of miRNAs in this context was a study by Triboulet et al demonstrating that two cellular miRNAs, miR-17-5p and miR-20 are down-regulated during HIV infection and target the histone acetylase PCAF24. PCAF is a co-factor to the Tat transactivator protein in HIV-1 and Triboulet et al showed that miRNA-mediated repression of PCAF suppresses HIV-1 production24. It is possible that downregulation of these miRNAs is therefore a viral mechanism for boosting the levels of proteins required for replication and spread. Further studies on the factors mediating miRNA down-regulation will shed light on this. Along this same theme, miR-100 and miR-101 were found to be down-regulated during human Cytomegalovirus (HCMV) infection and the targets of these miRNAs include genes in the mTOR pathway25, which is required for translation and replication of the virus26. Over-expression of miR-100 and miR-101 results in a 10 fold reduction in viral titre in human cells, suggesting that supplementing the levels of these miRNAs could be a therapeutic strategy for suppressing HCMV replication. Many other cellular miRNAs have now been linked to different viruses, based on their impact on replication and/or changes in their expression levels upon infection. For example, miR-23b is a RIG-1 dependent miRNA that downregulates VLDLR, a receptor required for viral entry of the Rhinovirus 1B (RV1B): overexpression of this miRNA prior to infection significantly reduces RV1B infection levels27. In contrast, miR-141 is induced by infection of enterovirus 71, another picornavirus, but this miRNA is advantageous to the virus as it downregulates EIF4E: this is a translational initiation factor whose down regulation helps shut-off host capdependent translation, thereby favouring cap-independent translation used by the virus28. Clearly, there is extensive diversity in how miRNAs can indirectly influence viral life cycles; a further list of miRNAs validated to display anti- or proviral properties by regulation of host genes is provided in Table 2. However, the interpretation of miRNA functions in a viral context merits careful consideration of cell type, stage of infection and interplay with viral pathogenesis. For example, increasing evidence suggests that control of cellular miRNA levels is a mechanism used by diverse viruses during latency29,30 and oncogenic viruses also deregulate miRNA implicated in carcinogenesis29,31-35. In addition, Kaposi sarcoma-associated herpesvirus (KSHV) infection deregulates miRNAs associated with cell motility and angiogenesis36. miRNA therapeutic strategies will therefore not only be relevant in controlling viral replication but also in controlling pathogenesis linked to cancer and other diseases.
TABLE 2 Indirect effects of miRNAs on viruses
| Viral family / Human pathogen | Cellular microRNAs | Function | Reference | |
| Herpesviridae | ||||
| HCMV | miR-199a-3p | Multipe genes | Antiviral | 41 |
| HSV-1 | miR-199a-3p | Multiple genes | Antiviral | 41 |
| KSHV | miR-132 | p300 | Proviral | 49 |
| Hepadnaviridae | ||||
| HBV | miR-155 | SOCS1 | Antiviral | 50 |
| Picornaviridae | ||||
| RV1B | miR-23b | VLDLR | Antiviral | 27 |
| EV71 | miR-141 | EIF4E | Proviral | 28 |
| Retroviridae | ||||
| HIV-1 | miR-17-5p, miR-20 | PCAF | Antiviral | 24 |
| Rhabdoviridae | ||||
| VSV | miR-155 | SOCS1 | Antiviral | 51 |
Although the above examples demonstrate rapid and important progress in this field, the knowledge of exactly how miRNAs exert their antiviral properties is still lacking. Most reports mention only one or two host targets of a miRNA. However, miRNAs can simultaneously regulate hundreds of proteins37,38 and can exert their functions by targeting host networks rather than individual genes39,40. It is also difficult to distinguish whether an antiviral effect of a miRNA is based on an interaction with the virus rather than the regulation of host genes (and presumably, there might be competition). We previously developed a global functional screening approach to identify cellular miRNAs with the more broad-spectrum antiviral properties41. We found that miR-199a-3p significantly reduced viral load in murine and human CMV, herpes simplex virus-1 and mouse gammaherpesvirus and the antiviral properties extended to Semlikiforest virus, an unrelated Togavirus41. Since the antiviral properties of miR- 199a-3p were conserved in these diverse viruses, it seems likely that the functional consequences relate to targeting host genes, rather than direct interactions with viral elements. Global mRNA analysis in cells treated with miR-199a-3p mimics versus inhibitors identified several hundred genes regulated by this miRNA, with a specific enrichment for pathways activated and required by many viruses: including ERK/MAPK signalling, PI3K/AKT signalling, prostaglandin synthesis, oxidative stress signalling and viral entry41. Although much work is still required to build a systematic model for how all of the targets of a miRNA collectively impact the cell, it is likely that the miRNA mechanism – the subtle downregulation of specific combinations of genes simultaneously – could hold potential for controlling diverse infections with less scope for resistance. This might also be a useful complement to existing antivirals. As with any host-targeted therapy, the effect of miRNA manipulation on a host cell will need to be carefully tested to ensure there are no adverse side effects.
The initial success with miR-122 inhibitors in HCV infection clearly gives weight to the utility of developing miRNA-based therapies as antivirals. In addition, compelling evidence for the importance of individual miRNAs in viral infection comes from recent studies showing that viruses have evolved mechanisms to specifically inhibit them. Cazalla et al recently showed that Herpesvirus saimiri (HVS) encodes a small RNA that specifically inhibits a cellular miRNA, miR-2742. This miRNA was also found to reduce replication capacity of murine CMV (distantly related to HVS) and MCMV also encodes an RNA that targets miR-27 for degradation (A. Buck, unpublished data)43. The fact that diverse viruses devote genome space to inhibiting specific cellular miRNAs is strong evidence for the importance of these host molecules in infection and the potential utility of developing drugs to EXPLOIT them.
About the Author
Amy Buck earned her PhD in Biochemistry at the University of Colorado in Professor Norman Pace’s lab where she studied RNA-protein dynamics. Wanting to apply her RNA background to the field of virology, she joined the Division of Pathway Medicine and the Centre for Infectious Diseases at the University of Edinburgh in 2005 and was awarded a Marie Curie Fellowship to study microRNA function and biogenesis in murine cytomegalovirus. In 2009, she was awarded an Advanced Fellowship through the Centre for Infection, Immunity and Evolution where she has built a research group focused on microRNA functions in host-pathogen interactions and mechanisms of microRNA regulation. For further information see www.bucklab.org.
Email: [email protected]