Automated workflow optimisation and assay development strategies for High Content Research Facility, Trinity College Dublin
Posted: 23 November 2007 | Dr Anthony Mitchell Davies, Institute of Molecular Medicine, Trinity College Dublin | No comments yet
High Content Screening (HCS) is becoming increasingly utilised as an early drug-discovery and basic research tool for defining the functions of genes, proteins and other biomolecules in normal and abnormal cellular functions. HCS involves the integration of a number of preparation steps which include; cell-sample preparation, fluorescent labelling, image acquisition, image processing, image analysis, information management and knowledge mining.
High Content Screening (HCS) is becoming increasingly utilised as an early drug-discovery and basic research tool for defining the functions of genes, proteins and other biomolecules in normal and abnormal cellular functions. HCS involves the integration of a number of preparation steps which include; cell-sample preparation, fluorescent labelling, image acquisition, image processing, image analysis, information management and knowledge mining.
High Content Screening (HCS) is becoming increasingly utilised as an early drug-discovery and basic research tool for defining the functions of genes, proteins and other biomolecules in normal and abnormal cellular functions. HCS involves the integration of a number of preparation steps which include; cell-sample preparation, fluorescent labelling, image acquisition, image processing, image analysis, information management and knowledge mining1.
These advanced microscopy and image analysis platforms are proving to be immensely powerful tools for the study of molecular and morphological events in cells. This allows for the first time, a multi-parametric characterisation of gross cellular responses and behaviour of a variety of molecular and cellular targets, including subcellular localisation and redistribution of individual proteins and complex cellular structures. In contrast to the more traditional biochemical or genetic analysis, HCS allows for the detection of physiological responses within the context of the structural and functional networks of cells in both normal and diseased states2.
High content screening facility, Trinity College
The High Content Research Facility was initially set up in 2004 as part of a larger strategic plan for Trinity Colleges’ Institute of Molecular Medicine. Since its inception, the utilisation of this research facility has grown to a point where it now plays a central role in numerous research projects and collaborations which take place in both the national and international scientific arenas. Over the last three years the reputation of this facility has grown to a point where it is now considered to be an international centre of excellence for both technical expertise and advanced training. The success of this facility has been underpinned by the efforts of our dedicated team of scientific specialists, whose primary research goal is the advancement of these technologies and their applications. We currently service approximately 30 research projects, which range from large scale gene silencing (siRNA) screens, where the functional characteristics of each individual gene in the whole human genome is assessed by disrupting its function. As well as gene silencing we are strongly committed in the area of nano-biology, where our facility is being currently utilised for study of the biocompatibility of a range of advanced nano-materials which have potential therapeutic and diagnostic applications. We also have numerous projects in the areas of infection and immunity, cell signaling, lung, gut and prostate cancer, heart disease, drug discovery and applied computing. Finally we have recently announced the successful completion of the worlds’ first academic course in High Content Screening and Analysis, where approximately 30 postgraduate students from a range of scientific and medical disciplines received the very latest theoretical and practical training from a host of world leading experts from industry and academia.
Technology profile and work flow
The High Content Research Facility at Trinity College Dublin is divided into distinct subsections, which include assay development, screening and high resolution con-focal imaging.
The high content screening process can be divided into three distinct stages. The first stage of the high content screening process is assay development. This process involves the optimisation of the cell based assay and HCS instrumentation. Typically this will involve optimisation of cell culture conditions (such as cell density and passage number), florescent stains, reporters and the measurable dynamic range of the cell based assay. This process is usually completed when a satisfactory Z factor for the cell based has been achieved. Much of our assay development work has been conduced using a Cellomics Kineticscan (Figure1), which is a HCS reader specifically designed for assay development.


Figure 1: Cellomics kineticscan HCS reader
The second stage in the high content screening process involves scaling up the cell based assay. In order to attain adequate throughput and high level of reproducibility, it is essential to translate these assays into an automatable format. This involves automating the various elements of the workflow. Much of the preparation, treating, fixing and staining of cells can be achieved using liquid handling systems. Our facility currently has two types of liquid handling robot which are used to complete these tasks. The first is the Matrix Hydra robot, which is a fully automated computer controlled system, fitted with a 96 well head that can dispense liquids directly into 96 or 384 well plates. The Hydra (Figure 2) can aspirate and dispense volumes ranging from 1 – 580 micro litres. This system also has a fully automated 3 position moving stage and a dedicated active wash bath. The hydra’s main functions include; bulk aspirating and dispensing of liquids (such as tissue culture medium, test compounds and fixing agents and stains), plate replication, plate washing, reformatting (96 to 384 and vice versa) and in well reagent mixing. The second system used in our labs is the Deerac Equator (Figure 3). This is an 8 channel fully automated liquid delivery system which has a dispensing range of 50nl-1ml per channel. This system is compatible with 96,384,1536 well plates. The equators main functions include; nano spotting, cell dispensing and its specialised gradient function makes it an ideal tool for setting up dose response and dye titration experiments.


Figure 2: The matrix hydra


Figure 3: The deerac equator
The third stage of the screening process is the image acquisition. This occurs at the point where cells are ready to analyse, in other words, the cells have been treated with test compounds or siRNA’s etc and appropriately stained. This stage of the process is conducted using the labs main High Content Screening instrument which is the INCELL 1000 (Figure 4). This High Content Screening system has also been fitted with a robotic plate feeder (Kinedx) and bar code reader with a 90 plate capacity. This instrument is capable of acquiring high resolution microscope images from 1-4 individual florescent dye channels (Figure 5). Once images are captured they are transferred to a high capacity server, where they are stored and subsequently analysed.


Figure 4: The Incell 1000
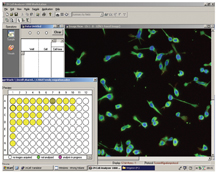

Figure 5: Screen shot of cellular images taken from Incell 1000
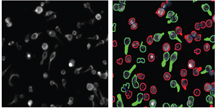
The forth stage in the high content screening process is image and data analysis. Image analysis involves the use of specialised algorithms, which have been designed to recognise whole cells and/or sub-cellular structures. This HCS image analysis software initially segments individual cells and/or sub-cellular structures (depending on the specific labelling of the cells), allowing the user to subdivide each cell into sub-cellular compartments such as nucleus and cytoplasm (Figure 6). Once segmentation has been achieved, the algorithm can extract information such as staining intensity and texture, and morphology such as shape and area from these defined sub-cellular regions. Another feature of most HCS image analysis packages is subpopulation analysis. This is an immensely useful tool if you wish to analyse sub sets of cells exhibiting a distinct biology; for example, measuring responses in transfected cells only whilst excluding data from un-tranfected cells in the same well. Subpopulation analysis has been useful in our hands for the measurement of cellular responses in migrating verses non migrating lymphocytes (Figure 7).
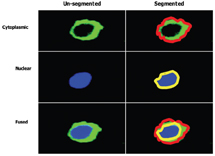

Figure 6: Illustrating a method of imaged based compartmentalisation of a single cell, which has been divided into two distinct spatial regions, (i) cytoplasmic (bounded by red line) and (ii) nuclear (bounded by yellow line). (Images supplied by Navin Verma Dept Clinical Medicine Trinity College Dublin)

The final step is analysis of the resulting data produced from the image analysis. Some HCS image analysis packages can generate anything up to 300 individual parameters on a per cell basis. Because of the huge amount of data produced, it is necessary to use a good data visualisation and analysis package such as Spotfire. Some of the HCS equipment vendors produce their own data visualisation package such as the Cellomics view package (Figure 8).
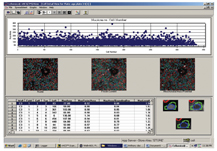

Figure 8: Screen shot of Cellomics data visualisation software. Used for first line data analysis
The fifth and final stage of our HCS process is a more in-depth analysis of any hits, which may have been identified. Analysis at this stage can take many forms and largely depends on the biology under study. However, as part of the Trinity HCS facility we have two high resolution con-focal microscope systems for more in-depth analysis. Con-focal analysis can be used to confirm localisation of target proteins that would occur during nuclear translocation. Additionally con-focal imaging can give precise information about the physical interactions of cellular proteins (Figure 9).
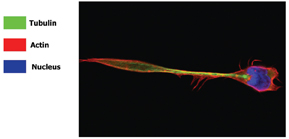

Figure 9: Con-focal image of a migrating lymphocyte, stained with Hoechst 33258 (nucleus), Phalloidin (actin cyto-skeleton) and anti-beta tubulin (tubulin cyto-skeleton). The localisation of the actin and tubulin, can be clearly seen from this image. (Image supplied by Navin Verma and Jenifer conroy, Dept Clinical Medince Trinity College Dublin)
Two selected projects currently being undertaken at the High Content Research Facility at Trinity College Dublin
Identification of the genes involved in the regulation of cell migration in T lymphocytes (Principle investigator: Dr Aideen Long (Senior Lecturer))
As part of an effective immune system, leukocytes continuously migrate from the circulation across the endothelium into tissue and interchange between the bloodstream and the lymphatics. In doing so they can patrol the body for antigen and enter sites of inflammation. This process is characterised by initial tethering of the leukocyte to the surface of specialised endothelial cells mediated largely by selectin interaction with their carbohydrate ligands expressed on the endothelial surface3,4. This slows leukocyte flow and cells roll along the surface of the endothelium.
Chemokines, secreted by cells within the tissue and immobilised on the endothelial cell surface can activate molecules of the integrin family on the rolling cells, leading to leukocyte arrest and subsequent diapedesis across the endothelium5,6. This process of extravasation is facilitated by polarisation of the cell, which crawls and squeezes between endothelial cells. Leukocytes polarise both in terms of shape and subcellular expression of receptors, adhesion and signaling molecules and cytoskeletal elements7,8. Actin, which is distributed throughout the lymphocyte, concentrates at the leading edge and along with microtubules distribute to the trailing edge or uropod (Figure 10).
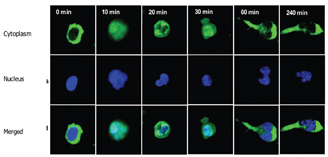

Figure 10: Two Colour images of T lymphocytes at various stages of the migration process, note at 60 minutes uropod is clearly visible. NB cytoplasm stained in green and nucleus in blue. (Images supplied by Navin Verma, Dept clinical medicine Trinity college Dublin)
Migratory signaling through integrins
Integrins are a widely expressed family of non-covalently linked · and ‚ subunits that mediate cell-cell and cell-extracellular matrix contacts. The integrins on circulating leukocytes bind minimally to their ligands but their adhesive capacity can be increased by stimulation of intracellular signaling pathways, for example, through the T cell receptor or by chemokines6. This mechanism of activation is termed inside-out signaling. Integrins may also be activated by divalent cations such as Mg2+ and Mn2+ or special stimulatory anti-integrin monoclonal antibodies. Active β1 and β2 integrins play an important role in extravasation of leukocytes from the circulation to sites of inflammation and in the homing of lymphocytes to secondary lymphoid organs.
Our cell based assay for study of T lymphocyte migration in HCS format
We have developed an in vitro model of T lymphocyte migration9,10. In this system, activated human T lymphocytes (PBTL) or the HuT 78 T cell line migrate (as demonstrated by time-lapse microscopy) when incubated with either immobilised recombinant ICAM-1-Fc fusion protein (the natural ligand for the leukocyte function-associated antigen-1 (LFA-1)) or a triggering antibody to the αL-chain of the LFA-1 molecule itself. T lymphocytes stimulated through the LFA-1 molecule adopt a locomotion-associated polarised phenotype. With this migratory phenotype the cell acquires a long uropod or ‘tail’ which can be several times the actual diameter of the cell body. Upon further analysis of the polarised cells in this model, it was demonstrated that the microtubule cytoskeleton undergoes a dramatic rearrangement and is distributed primarily in the trailing uropod of migrating cells. Expression of the enzyme Protein Kinase Cβ, which associates with the centrosome and microtubule cytoskeleton in migrating T cells is necessary for this process of T cell polarisation and migration11.
As T cell migration is central to the inflammatory process, we believe that our model of T cell migration is ideal for use in a screening environment for the study of the mechanism(s) of migration (for example, the elucidation of signaling/regulatory molecules involved) and for the screening of potential drugs/anti-inflammatory compounds. To this end we have adapted the T cell migration assay to a multi-well format for use in High Content Analysis (HCA). In this system, plates are coated with antibodies, which stimulate T cell migration and following polarisation cells are fixed and stained with cytoskeletal markers such as phalloidin (labels actin). The shape of the cytoskeleton is reflective of the migratory capacity of the cell and HCA algorithms such as those which calculate cell elongation or cell area can be used to distinguish round (non-migratory) cells from polarised (migratory) and in some cases hyperpolarised cells. We propose to use this system to screen siRNA libraries and small molecule/ pharmacological compounds to interrogate the mechanisms involved in the migratory process and thus potential pro-inflammatory indicators. These screens will be performed in a high throughput environment utilising, for example, 96-well electroporation device (for delivery of siRNA) and the liquid handling facilities for dispensing of cells and reagents for staining (Figure 11).
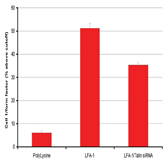

Figure 11: Showing results of experiments performed, to determine the effects of siRNA knockdown of talin, which is known to be involved in cell migration. Results expressed as 1/form factor (GE Healthcare HCS Investigator image analysis Package) Migration was measured in cell seeded on Poly-Lysine and LFA-1 which are know to inhibit and promote cell migration, respectively).
Development of cell based assay tools for the study of cardiac disease and cardiotoxicty
Cardiotoxicity and drug discovery
Drug discovery and development is a complex process which consumes vast financial and technical resources (average investment before market $897 Million over a 12 year Period). This large investment is partly due to high attrition rates, approx 90% of candidate compounds fail and approx 50% lost due to toxicity and poor efficacy.
Many compounds fail later on in the discovery process resulting in increased costs to the drug discovery industry. Potential cardiotoxicity is an ever present factor even when drug reaches market. Examples of drug market withdrawals; Seldane (terfenadine) Antihistamine, Raxar (Antibiotic) and COX2 Inhibitors such as Bextra (Valdecoxib). Drug discovery organisations are attempting to reduce costs by implementing fast to fail strategies but as yet there is no effective cell based assay or technology which can be utilised for early detection of potential cardio-toxicity.
The social and economic costs of heart disease
Cardiovascular disease accounts for approximately 33% of all annual mortalities globally and about 43% in the Western World. Heart disease is presenting earlier in ever increasing numbers year on year. It has been estimated that by 2030, heart disease and associated conditions such as stroke will become the leading cause of death and disability world wide, rising to approximately 24 million a year.
The problem
With the exception of some automated electrophysiological assays such as those for IKR channel inhibition (hERG) there very few reliable cell based assays which can be utilised for early pre-clinical cardiac safety or efficacy screens. The heart is a very complex organ consisting of many different cell types, to ensure effective cardiac function all cells must work in a highly co-ordinated fashion. Any of these cells may be a potential therapeutic target or site of a toxic event.
Our studies are focused on the ventricular myocytes, as these are the main effector cells in the heart. It is the ventricular myocytes that generate the force which drives the blood around the circulation. Ultimately a drugs cardiac efficacy or safety will be judged by how it influences the hearts ability to generate this hydrostatic pressure (perform as a pump). Unfortunately such functional parameters are difficult to measure at the cellular level in vitro. Ideally we need a range of assays which will allow us to study functional characteristics of the ventricular myocardium at the cellular level in an automateable format, at a level of throughput which will be useful to drug discovery organisations.
Considerations for setting up an cardiac cell based assay
As with any type of cell based assay, it is desirable to set up an assay which reflects the biology understudy. This is especially important when studying the behaviour of cardiac cells in vitro. Firstly it is important to choose a cell type which is physiologically relevant and appropriate for the assay requirements. For example, it is widely believed that primary adult cardiomyocytes are more biologically relevant than using isolated organelles or engineered cell lines. This is because the cyto-skeleton and signaling networks remain intact and functional. In addition, intact cell functionality allows for a more realistic assessment of cellular responses to candidate drugs and so on. However there are problems associated with using primary cardiac cells and tissues. These include animal maintenance and difficulties isolating these cells. In addition, they are only viable for a few days and are highly sensitive to mechanical damage. Despite these disadvantages, we believe that these cells are ideal for use in functional assays and can be used in secondary or tertiary screens. To get the most reliable data from these cells, attempts have been made to mimic the physiological and environmental conditions that these cells are exposed to in the heart (Figure 12).
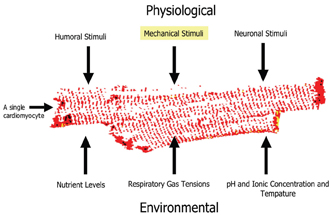

Figure 12: Showing some of the key physiological and environmental factors which should be considered when setting up a cell based assay using isolated adult cardiomyocytes.
As can be seen from Figure 12, there are numerous factors that need to be considered when setting up such a cell based assay. So far our studies have been focused on mechanical stimuli, which we can reproduce by inducing the myocytes to contract in response to an applied electrical impulse. We are working on the theory that the contraction dynamics of cardiac myocytes, for example, rate of contraction (cell shortening) and relaxation12, can be viewed as an integration of many of the key cellular process which occur in a contracting myocyte. Likewise the contractile activity of a given myocyte will also result in changes on those same cellular processes (Figure 13). An example of where mechanical activity can influence cellular state is shown in Figure 14, which demonstrates typical survival times of either quiescent or contracting primary adult cardiac myocytes maintained at different oxygen tensions.
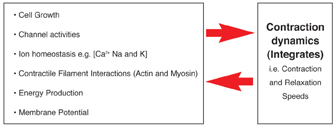

Figure 13: Showing the inter-relationship between contraction dynamics of cardiomyocytes and key sub-cellular processes, such as cell metabolism, ion homeostasis etc
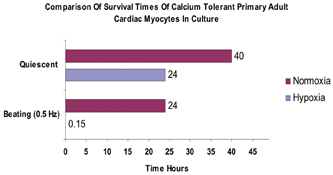

Figure 14: Showing the typical survival times (hours) of quiescent and beating adult cardiomyocytes maintained in either a normal oxygen atmosphere (atmospheric 21%) or in reduced oxygen ( hypoxia) < 5% ) oxygen.
As can be seen from Figure 14, both quiescent and contracting cells survive between 24-48 hours in normoxic conditions. In hypoxic conditions, quiescent cells can still survive for approximately 24 hours. However, beating cells maintained in hypoxic conditions tend to survive for approximately 10-20 minutes, thus demonstrating that the mechanical activity of cardiomyocytes has a direct effect on their survivability in culture. Indeed in vivo, cardiac cells would only normally be expected to survive for a few minutes in hypoxic conditions, thus suggesting that this beating cell model, will offer a more physiologically relevant cell based assay than using quiescent cardiomyocytes alone. We are currently developing a 96 well based system for electrically stimulating cardiac cells. This will allow us to conduct large scale experiments for assessing the effects of drugs on cardiac cells in a high content automatable format. Finally, we have been evaluating our high content screening systems ability to discriminate between living cardiomyocytes and those which are dead or dying (Figure 15). As can be seen from Figure 15, the image analysis software can clearly discriminate between the healthy rod shaped calcium tolerant cells (bounded by green) and the unhealthy hyper-contracted calcium intolerant short rounded cells (bounded in red).
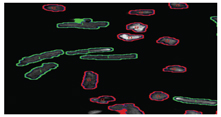

Figure 15: Showing screen shot of a mixed population of calcium tolerant (rod shaped bounded in green) and calcium intolerant primary adult cardiomyocytes (short and rounded bounded in red).
Finally; we have been assessing the suitability the H9C213,14 cell line, which is a cardiac derived myoblasic rat cell line, for a primary high content screen. These cells offer a number of advantages over the primary cells discussed earlier, for example; no complicated isolation or culturing procedures required, stable in culture for many days after seeding. In addition, these cells have functional RYR and SERCA, as well as intact Alpha and Beta adrenoreceptor signaling pathways.
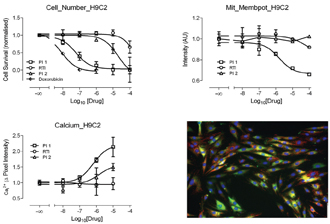

Figure 16: Showing results from cells were treated with 3 known cardio-toxic agents 2 protease inhibitors (PI 1 and 2) and 1 reverse transcriptase inhibitor (RTI). Measurements were taken from cells which were triple stained wit Fluo 4 (green). TMRM (red) and Hoechst (blue). The micrograph bottom right hand side is a fused image of triple stained cells.
We are currently using these cells to examine the toxic effects of a small panel of anti-viral drugs which are known to be cardio-toxic. In this study, cells were treated with 3 known cardio-toxic agents 2 protease inhibitors (PI 1 &2) and 1 reverse transcriptase inhibitor (RTI). From previous studies, it was indicated that the PIs would have a stronger effect than the RTI. Measurements were taken from cells which were triple stained wit Fluo 4; TMRM and Hoechst, to enable measurements of intracellular calcium, mitochondrial membrane potential and nuclear morphology (respectively), as can be seen from Figure 16. As anticipated the PIs seem to exert a greater toxic effect than the RTI. This is reflected in a dose dependant decrease in cell number and mitochondrial membrane potential accompanied by an increase in intracellular calcium.
In conclusion
HCS is allowing us to conduct information rich cell based experiments, in both live and fixed endpoint assays. The automatability of these assays and relatively small sample sizes allows us to conduct large scale experiments, with short turn around times and at low cost. Without these systems much of the work described earlier would be impossible to conduct at the scale required. It is our opinion that HCS technology will become a standard technology in most academic research institutions within the next five years.
Acknowledgments
- Professor Dermot Kelleher (Dean Trinity Medical Faculty)
- Dr Aideen Long (Senior Lecturer) Principle investigator Cell migration
- Dr Yuri Volkov (Senior Lecturer) Principle investigator Nano technology
- Dr Southeng Ong (Cell Signaling Assay Development)
- Dr Paul Spiers Pharmacology
- Dr Dara Dunican (Cell migration screen-senior assay development)
- Dr Emily Bennet (Cell Migration Screen Senior scientist)
- Navin Verma (Cell signaling/cell migration)
- Jennifer Conroy (Confocal Facility)
- Connla Edwards (High Content Research)
- Ronan Doyle (High Content Research)
References
- Korn K, Krausz E. Cell-based high-content screening of small-molecule libraries. Curr Opin Chem Biol. 2007 Oct 9;
- Abraham VC, Taylor DL, Haskins JR. High content screening applied to large-scale cell biology. Trends Biotechnol. 2004 Jan;22(1):15-22. Links
- Chen M, Geng JG. P-selectin mediates adhesion of leukocytes, platelets, and cancer cells in inflammation, thrombosis, and cancer growth and metastasis. Arch Immunol Ther Exp (Warsz). 2006 Mar-Apr; 54(2):75-84.
- Ley K, Reutershan J. Leucocyte-endothelial interactions in health and disease. Handb Exp Pharmacol. 2006 ;(176 Pt 2):97-133.
- Alon R, Dustin ML. Force as a facilitator of integrin conformational changes during leukocyte arrest on blood vessels and antigen-presenting cells. Immunity. 2007 Jan; 26(1):17-27.
- Kellersch B, Kolanus W. Membrane-proximal signaling events in beta-2 integrin activation. Results Probl Cell Differ. 2006;43:245-57
- Serrador JM, Vicente-Manzanares M, Calvo J, Barreiro O, Montoya MC, Schwartz-Albiez R, Furthmayr H, Lozano F, Sanchez-Madrid F. A novel serine-rich motif in the intercellular adhesion molecule 3 is critical for its ezrin/radixin/moesin-directed subcellular targeting. J Biol Chem. 2002 Mar 22;277(12):10400-9.
- Millan J, Montoya MC, Sancho D, Sanchez-Madrid F, Alonso MA. Lipid rafts mediate biosynthetic transport to the T lymphocyte uropod subdomain and are necessary for uropod integrity and function. Blood. 2002 Feb 1;99(3):978-84.
- Volkov Y, Long A, Kelleher D. Inside the crawling T cell: leukocyte function-associated antigen-1 cross-linking is associated with microtubule-directed translocation of protein kinase C isoenzymes beta(I) and delta. J Immunol. 1998 Dec 15;161(12):6487-95.
- Long A, Mitchell S, Kashanin D, Williams V, Prina Mello A, Shvets I, Kelleher D, Volkov Y. A multidisciplinary approach to the study of T cell migration. Ann N Y Acad Sci. 2004 Dec;1028:313-9.
- Volkov Y, Long A, McGrath S, Ni Eidhin D, Kelleher D. Crucial importance of PKC-beta(I) in LFA-1-mediated locomotion of activated T cells. Nat Immunol. 2001 Jun;2(6):508-14.
- Bers, D.M. 2001. Excitation-Contraction Coupling and Cardiac Contractile Force 2nd ed., Developments in Cardiovascular Medicine, Vol. 237. Dobson, Jon. 2007. Pediatric Respiratory Reviews Volt 8, Issue 1, March 2007, Pages, 62-66
- Hatcheller, J et al. 199. Morphological, biochemical, and electrophysiological characterisation of a clonal cell (H9c2) line from rat heart Circ. Res. 1991;69;1476-1486. Kimes, B. W., & B. L. Brandt. 1976. Properties of a clonal muscle cell line from rat heart. Exp. Cell Res. 98: 367-381
- Lee, H & Wei, Y. 2005. Mitochondrial biogenesis and mitochondrial DNA maintenance of mammalian cells under oxidative stress. Int J Biochem Cell Biol (2005) 37: 822-34.
Dr Anthony Mitchell Davies
Institute of Molecular Medicine, Trinity College Dublin
Dr Davies received his PhD in molecular Cardiology (BHF Funded) from the Faculty of Medicine, University of Dundee Scotland. This was followed by a research position (BHF Funded) which focused on the development of specialised cell based assays and perfusion systems for the study of hypoxic and ischemic insult in cardiac muscle cells and tissues (Tayside Institute of Child Health Dundee Scotland).
Dr Davies then undertook further research (Scottish Enterprise Funded) which focused on the development of cell based assays and living cellular arrays for high throughput and high content screening using adult primary cardiomyocytes. His current position is as director of high content research facility at Trinity College Dublin.




