Target validation
Posted: 20 May 2005 | | No comments yet
All diseases have a genetic component, whether inherited or resulting from the body’s response to environmental stresses such as viruses, toxins or trauma. The successes of the human genome project have enabled researchers to pinpoint errors in genes that cause or contribute to disease.
All diseases have a genetic component, whether inherited or resulting from the body's response to environmental stresses such as viruses, toxins or trauma. The successes of the human genome project have enabled researchers to pinpoint errors in genes that cause or contribute to disease.
All diseases have a genetic component, whether inherited or resulting from the body’s response to environmental stresses such as viruses, toxins or trauma. The successes of the human genome project have enabled researchers to pinpoint errors in genes that cause or contribute to disease.
The crucial final stage in validation of novel treatment target is the in vivo confirmation of the role of the gene in a disease model in order to link the gene with a disease. At this moment, the only available method for such validation is to create a mouse that lacks the gene of interest (knock-out mouse) and thereby demonstrate the role of that gene in disease. Despite continuous successes using this approach, this method has several drawbacks. It is still extremely time-consuming, rather costly and has the major drawback that genes beside their role in disease, also play a general role in the body that cannot be studied (removal of such gene will cause death), unless conditional or local knock-outs are generated. Also for every gene of interest a novel mouse must be generated. Therefore other technologies are needed that complement the existing knock out mice in target validation studies. Initially antisense oligonucleotides were used, but now there is a new kid on the block: RNA interference (RNAi). With the rise of the RNA interference (RNAi) technologies, steps are being made to develop such alternatives, using an approach to knock-down gene function (inactivating the gene at an mRNA level rather than elimination of the gene on a DNA level; also known as post-transcriptional gene silencing).
RNA interference
RNAi is a molecular mechanism that evolved during evolution to protect cells against invasion of foreign genes and was first described in Caenorhabditis elegans1. Since then the mechanism has been demonstrated in a variety of eukaryotes such as insects, plants, funghi and vertebrates2. This protective mechanism is now exploited to specifically knock down the function of a gene in both mechanistic studies as well as in therapeutic approaches. RNAi is the induction of sequence specific gene silencing by double stranded RNA molecules. In vivo, long dsRNA molecules are processed by Dicer (a member of the RNase III family of ribonucleases) into the small interference RNA (siRNA) molecules of 21-23 nucleotides in length. These siRNA molecules subsequently bind to an enzyme complex known as RISC (RNA induced silencing complex), which unwinds the dsRNA, facilitates base pairing to the specific target mRNA and cleaves the mRNA, thus silencing gene expression. Since in mammalian cells the long dsRNA precursor molecules also induce interferon responses that change cellular behaviour, these long dsRNA molecules are not used in mammalian cells. Rather, short siRNA molecules are used, thereby circumventing Dicer and the interferon response, whilst retaining effective destruction of the targeted mRNA.
In vivo requirements
Ample evidence is available on the applicability of siRNA for target validation using in vitro cell systems. The current challenge is to apply this target validation tool in vivo in preclinical animal models as well, but relatively few reports have been published in this area. Most of these studies show proof-of-principle studies in which an exogenously added reporter gene is knocked-down by co-transfection of siRNA against the reporter gene. For example, intravenous injection of cationic liposomes complexed to a plasmid carrying a GFP together with antiGFP siRNA, resulted in significant reduction of GFP production both on mRNA as well as on protein level in liver and spleen3. A limited – but growing – number of studies show effective knock down of an endogenous gene resulting in a functional change in vivo. In general, in vivo studies are hampered by some of the same problems that also complicate in vivo use of antisense oligonucleotides or gene therapy approaches. The hurdles that need to be tackled include the effective and specific delivery of siRNAs into the target cells or tissue and the duration and efficacy of gene knock-down.
Synthetic siRNA
Double stranded siRNA molecules are more stable than single stranded antisense oligonucleotides, which should facilitate their in vivo applicability, but delivery of short, naked, synthetic siRNA molecules by intravenous injection also leads to rapid degradation of the siRNA molecules. Still, in short-term models the injection of naked siRNA could be useful. For example, intraperitoneal injection of anti-TNFα siRNA in mice reduced peritoneal TNFα levels (but not IL-1α levels, indicating specificity) and concomitantly protected against injection (18 hours after the siRNA delivery) of a lethal dose of LPS4. High pressure injection of plasmids into tail vene results in efficient uptake of a plasmid in the liver. Similarly, high volume injection of siRNA in the murine portal vene results in transfection of 60 per cent of the hepatocytes. Blockage of caspase-8 or caspase-3 production using this method rescued 30-50% of mice that would normally die following surgically induced total hepatic ischemia5.
For longer stability, several types of chemically modified siRNA molecules have been used and this area is still evolving rapidly, similar to the chemical modification approaches that have been tested for antisense oligonucleotides in the past. siRNA containing phosphorothioate exibit increased serum stability6. Modifications to the sugar moiety of the siRNA molecule such as 2’-O-methyl ribonucleotides or RNA analogues with a methylene linkage between the 2’and 4’ positions enhance resistance to nuclease mediated degradation and improve thermal stability, while maintaining target knock down efficacy.
Also, carrier proteins have been used to stabilise the siRNA. Atelocollagen complexed with siRNA is resistant to nucleases and is efficiently transduced into cells, thereby allowing long-term gene silencing7. In an alternative approach noncovalent complexation of synthetic siRNA with low molecular weight polyethylenimine was used to deliver siRNAs (against a growth factor receptor) to subcutaneous tumors following intraperitoneally delivery in mice, resulting in a marked reduction in tumor growth8.
Plasmid-driven siRNA
To ensure prolonged presence of siRNA the logical step is to have the siRNA produced inside the target cell. Such local production of siRNA may be achieved by transfecting the cells with plasmids that produce short-hairpin RNAs (shRNAs, which are siRNA molecules that are synthesised as sense strand – hairpin – anti-sense strand that spontaneously form RNA duplexes) or siRNAs. The production can be driven by constitutive, inducible or cell specific promoters thereby creating a highly flexible system. Similar to the synthetic siRNA approaches described above, delivery of the plasmids may be facilitated by a variety of carriers, including lipid vehicles or by high volume injections. McCaffrey and co-workers used the later approach to co-transfect a plasmid expressing hepatitis B viral surface antigen (HBVsAg) together with a plasmid expressing an appropriate shRNA into hepatocytes of immuno-competent as well as immunodeficient mice. They show a reduction of more than 80 per cent of HBVsAg production on mRNA as well as protein level, for as long as one week after transfection9.
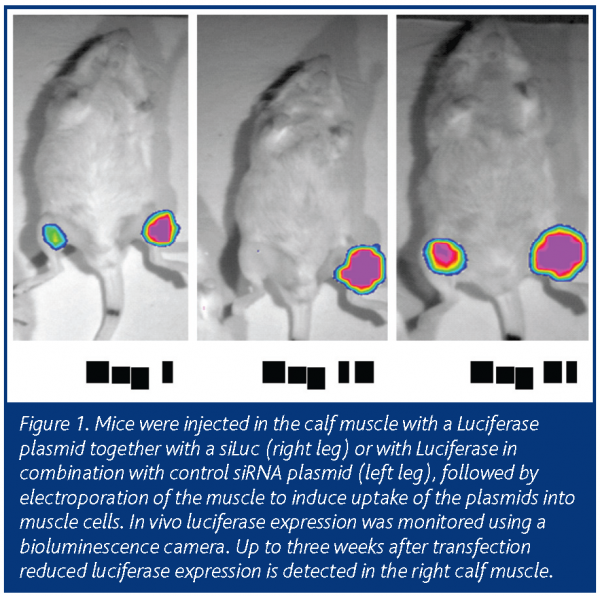
For target tissues other than the liver, other delivery methods are required. Electroporation-mediated gene transfer to the muscle of living animals is a very efficient way to obtain long-term expression of the introduced transgenes or siRNA constructs. Using this strategy, we were able to demonstrate the siRNA-mediated silencing of a co-transfected luciferase plasmid in vivo up to three weeks after transfection (Figure 1; unpublished data).
Viral delivery of siRNA
In addition to the above, viral delivery of siRNA is a powerful way to achieve prolonged gene knock-down in vivo. Several studies have been published using different types of viruses that target (either because of their trophism or because of the way they were injected) different tissues/cells. Injection of an adeno-associated viral vector delivered a shRNA against tyrosine hydroxylase (a key enzyme in the production of the neurotransmitter dopamine) into the brain of nine-week old mice significantly impaired locomotion coordination, mimicking features of Parkinson’s disease (which is characterised by the degeneration of dopaminergic neurons). This approach demonstrated the use of RNAi to generate models of disease and concomitantly points at a potential pivotal role for tyrosine hydroxylase in the development of Parkinsons disease10. Subretinal co-injection of adenoviruses producing enhanced green fluorescent protein and an appropriate siRNA that blocks production of this reporter protein shows that siRNA can knock down genes in retinal cells. Similar delivery of an siRNA directed at the murine vascular endothelial growth factor significantly reduces laser-induced neo-vascularization demonstrating potential for therapeutic as well as target validation applications in retinal disease11. Initial experiments using the rat collagen-induced arthritis model for rheumatoid arthritis showed that intra-articular delivery of an adenovirus producing antiTNF siRNA blocks disease development by ~30 per cent; similar to intravenous treatment with Remicade (an anti-human TNF antibody that is on the market for arthritis patients) (unpublished data). These data underscore the power of siRNA as target validation tool in animal models of disease. The advantage of these viral delivery methods is clearly the efficient delivery of nucleic acids to the target cell. Modification of viral proteins can be used to change trophism and thus target cell specificity. The drawback of all viral approaches is the potential side-effects that are induced by the viral proteins in an organism, such as the pro-inflammatory effects of certain adenoviruses.
Conclusions
RNAi is rapidly become one of the major tools in functional genomics studies in biology and biomedicine. The technology builds on the vast experience within the research community on antisense oligonucleotides and gene therapy techniques and also encounters some of the problems that these technologies face. The studies described above show that for RNAi to grow into its full potential it is crucial to further develop appropriate methods to bring the siRNA molecules into the right cell/tissue in vivo. Both viral and non-viral (such as electrotransduction) techniques will seem most promising in this respect.

Reference
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998; 391(6669):806-811.
- Milhavet O, Gary DS, Mattson MP. RNA interference in biology and medicine. Pharmacol Rev 2003; 55(4):629-648.
- McCaffrey AP, Meuse L, Pham TT, Conklin DS, Hannon GJ, Kay MA. RNA interference in adult mice. Nature 2002; 418(6893):38-39.
- Sorensen DR, Leirdal M, Sioud M. Gene silencing by systemic delivery of synthetic siRNAs in adult mice. J Mol Biol 2003; 327(4):761-766.
- Contreras JL, Vilatoba M, Eckstein C, Bilbao G, Anthony TJ, Eckhoff DE. Caspase-8 and caspase-3 small interfering RNA decreases ischemia/reperfusion injury to the liver in mice. Surgery 2004; 136(2):390-400.
- Braasch DA, Paroo Z, Constantinescu A, Ren G, Oz OK, Mason RP et al. Biodistribution of phosphodiester and phosphorothioate siRNA. Bioorg Med Chem Lett 2004; 14(5):1139-1143.
- Minakuchi Y, Takeshita F, Kosaka N, Sasaki H, Yamamoto Y, Kouno M et al. Atelocollagen-mediated synthetic small interfering RNA delivery for effective gene silencing in vitro and in vivo. Nucleic Acids Res 2004; 32(13):e109.
- Urban-Klein B, Werth S, Abuharbeid S, Czubayko F, Aigner A. RNAi-mediated gene-targeting through systemic application of polyethylenimine (PEI)-complexed siRNA in vivo. Gene Ther 2005; 12(5):461-466.
- McCaffrey AP, Nakai H, Pandey K, Huang Z, Salazar FH, Xu H et al. Inhibition of hepatitis B virus in mice by RNA interference. Nat Biotechnol 2003; 21(6):639-644.
- Hommel JD, Sears RM, Georgescu D, Simmons DL, DiLeone RJ. Local gene knockdown in the brain using viral-mediated RNA interference. Nat Med 2003; 9(12):1539-1544.
- Reich SJ, Fosnot J, Kuroki A, Tang W, Yang X, Maguire AM et al. Small interfering RNA (siRNA) targeting VEGF effectively inhibits ocular neovascularization in a mouse model. Mol Vis 2003; 9:210-216.



