Applying systems biology and computer simulations to predicting idiosyncratic DILI
Posted: 19 August 2010 |
Idiosyncratic drug-induced liver injury (DILI) is a rare adverse drug reaction which accounts for a significant amount of patient suffering, including death. Currently, idiosyncratic DILI is unpredictable and as a result arises late in the drug development process or even post-marketing. The prediction of idiosyncratic DILI based on preclinical or early clinical data is a formidable challenge and this short review will discuss why and how new initiatives in systems biology and dynamic computational simulations can meet this challenge and predict the ‘unpredictable’.
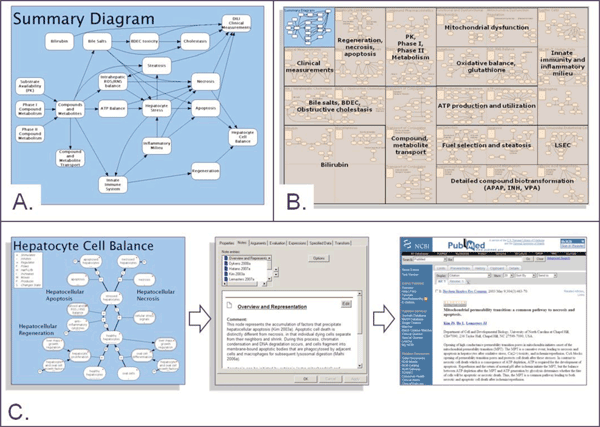

Figure 1 Overview of DILI-sim A. Schematic overview of the key biological processes represented in DILI-sim B. Overview of different modules within DILI-sim. Each module is itself a model that captures a specific area of relevant biology, pharmacology and metabolism. This modular approach to DILI-sim allows the overall model to be built in manageable, testable pieces C. Knowledge management aspects of DILI-sim. Under each of the models, the supporting evidence is explicitly captured and hence the model acts as a highly structured knowledge repository
Idiosyncratic drug-induced liver injury (DILI) is a rare adverse drug reaction which accounts for a significant amount of patient suffering, including death. Currently, idiosyncratic DILI is unpredictable and as a result arises late in the drug development process or even post-marketing. The prediction of idiosyncratic DILI based on preclinical or early clinical data is a formidable challenge and this short review will discuss why and how new initiatives in systems biology and dynamic computational simulations can meet this challenge and predict the ‘unpredictable’.
Drug-induced liver injury (DILI) is a potential undesirable complication for many drugs, which can result in life-threatening hepatotoxicity and subsequent patient mortality1. DILI can be classified as either ‘intrinsic’ (predictable and dose-dependent e.g. for acetaminophen) or ‘idiosyncratic’ (unpredictable and not necessarily dependent on dose). The current approaches taken to the identification of hazards and predictions of safety risk for new pharmaceuticals are relatively effective at identifying those with the potential to produce intrinsic DILI but are less effective in the early detection and prediction of idiosyncratic DILI and as a result, idiosyncratic DILI accounts for most of the hepatotoxicty observed in response to pharmaceutical use. Although the general prognosis for patients with DILI is generally good2, it still results in a significant amount of patient suffering and deaths annually. Additionally, the unpredictable nature of idiosyncratic DILI and the fact that it occurs in a minority of patients means that the detection of idiosyncratic DILI for new medicines tends to occur late in the development phase or even post-marketing and as a result, DILI has been the cause of several late-stage drug withdrawals costing the industry billions of dollars3.
The underlying mechanisms contributing to DILI are diverse and include drug-metabolism, pharmacology and biochemical drivers4. Additionally, there is evidence of a link between immune function (both innate and acquired) and the production of DILI1,4,5 and hence although the outcome may be hepatotoxicity, the triggers and drivers for it may lie in organ compartments different to and distal from the liver itself. The risk factors that contribute to idiosyncratic DILI are numerous and poorly understood. A recent review by Chalasani and Björnsson1 highlights age, sex, genetic background, drug dose, drug interactions, metabolic state, underlying comorbidities and alcohol as all potential risk factors contributing to DILI. There is no single factor that accounts for an increased risk of idiosyncratic DILI and sometimes the evidence is contradictory as to whether a factor is a risk or not. It is likely that the development of idiosyncratic DILI is therefore both multifactoral and temporal, requiring a ‘perfect storm’ of pharmaceutical, genetic, biological and environmental factors. It is for these reasons that current approaches to prediction of DILI often fail to detect or predict idiosyncratic potential; even patients within a large-scale clinical trial may not represent the appropriate ‘model’ for detection of an idiosyncratic adverse drug reaction. One consequence of this has been that regulatory agencies are demanding larger and longer clinical trials, beyond that required to establish efficacy, in order to try and establish drug safety. This increases costs and creates a bottleneck in the delivery of new medicines to the market.
The challenges in improving prediction of idiosyncratic DILI are formidable as they require techniques that can use data generated by preclinical screens or early clinical trials to predict the potential occurrence of DILI in a small number of patients within a broader patient population. In this short review, I will argue that systems biology approaches, specifically dynamic simulations, can meet this challenge and discuss some new initiatives that offer the potential to finally move idio – syncratic DILI from the realms of ‘unpredictable’ to ‘predictable’.
Systems biology
The discipline of ‘systems biology’ is one that has emerged over the last 10 years and is the application of computational and mathematical analysis to generate and test biological hypotheses. The central philosophy of systems biology is one that rejects the ‘reductionist paradigm’, in which biological systems are broken down into their elemental pieces, in favour of a more holistic approach to biology that attempts to put the vast amount of molecular information and data now available back into the context of the system from which it came. Essentially, systems biology can be thought of as “physiology with numbers” (Ian Wilson, personal communication) and at the heart of the discipline sits the application of a range of computational and mathematical techniques to the analysis of biological and pharmaceutical data and knowledge. Systems biologists take a quantitative view of biology which allows the more effective consideration of how different components and factors within any biological system can contribute to its overall behaviour. The different approaches used in systems biology are described by the terms ‘models and modelling’ but this simple phrase belies the diversity of different methods it encompasses. Despite the potential of systems biology to transform the drug discovery approach6, the discipline has struggled to find its niche as a standard approach in pharmaceutical research and development. Unlike the use of models in areas such as quantitative structure activity relationship (QSAR) analyses or drug metabolism and pharmacokinetics, the routine application of modelling to broader questions in biology is limited.
Models in systems biology can be roughly divided into ‘static’ and ‘dynamic’. Static models don’t include time as a variable and use statistical methods to find correlations within datasets. Examples of static modelling include QSAR analysis, OMIC (transcript-profiling, proteomics metabolonomics/metabonomics) analysis and network or pathway analysis. In contrast, dynamic models seek to describe how systems evolve over time and hence are often built using differential equations. Examples of dynamic models are those used to predict pharmacokinetic properties and biological simulations of biochemical pathways, organs and even whole organisms7,8,9. It is this latter category of models that may have the potential to improve the prediction of idiosyncratic DILI.
Although the mathematics within a model may appear impenetrable, a model is simply a tool with which to integrate a large amount of information. In effect, it is a summary of the knowledge and data within an area of biology and if this knowledge or data is incomplete or wrong, then the model will reflect this. To cover gaps in knowledge, models can also contain assumptions, but unlike the assumptions contained with the heads of scientists, those within a model have to be explicitly stated and are therefore open to challenge and debate. The very act of modelling a biological system can therefore be an exercise in summarising the knowledge and assumptions as to how the system behaves in a very transparent way and often this is the most valuable part of a modelling exercise.
Once constructed, models can be used to perform thousands if not millions of ‘in silico’ experiments. The effects of changes to all of the elements within the model can be systematically considered (alone or in combination) and those most important in dictating its behaviour identified. It is therefore possible to investigate how effects of differences between species will affect the behaviour of the system. Beyond this, and more importantly from the perspective of idiosyncratic DILI, the effects of strain and genetic variation within a population can also be considered and predictions made as to how these could impact on the system’s behaviour. The differences that such changes make to the model can be quantified and it is possible to predict not only what is important, but also what is not.
Prediction of idiosyncratic DILI is a systems biology problem
As discussed above, the development of idiosyncratic DILI within a patient is due to the combination of a number of factors triggered by the administration of a pharmaceutical. Patients who suffer idiosyncratic DILI responses do not have unusual biochemistry per se, but an unusual set of circumstances. The production of idiosyncratic DILI is therefore multifactorial (the integrated outcome of many factors), spatial (due to the interplay of different cell types and systems), temporal (occurring over time) and, by definition, only occurring in a minority of situations. Making an effective prediction of idiosyncratic DILI therefore demands taking a quantitative approach to analysis because as soon as there is a need to consider the combination of many factors, one must understand not only what contributes to DILI but also by how much. The only way to effectively approach the question of ‘how much’ is by using mathematical models because the sort of qualitative arguments often deployed by biologists are simply not sufficient. It is for this reason that the prediction of DILI is a problem in systems biology. To translate idiosyncratic DILI into the language of systems biology, idiosyncratic DILI is due to the pharmacological perturbation of an atypical biological system within a population of biological systems.
There is precedence for the use of systems modelling in the prediction of drug-induced toxicities in other organ systems7 and the development of models for the prediction of idiosyncratic DILI is now moving beyond the theoretical. Subramanian et al.10 describe the development of an organ-level systems model of liver. This model focuses on intrinsic processes in liver metabolism that can contribute to DILI including energy metabolism, glutathione metabolism, fatty acid and triglyceride metabolism and bile acid transport and metabolism. The model aims to simulate the ‘normal’ liver state and successfully represented the basic energetic state of a hepatocyte together with concentrations of some key intracellular metabolites. Using this model, it was possible to simulate the effects of several drugs and toxins on the liver with results that were in agreement with experimental measurements. The drugs and toxins considered in this publication are intrinsic hepatoxins but the authors also describe an in silico experiment in which they considered the range of hydrogen peroxide production that could be tolerated by their virtual liver cell. The results of this experiment were consistent with observations in genetic variations in superoxide dismutase (the generator of hydrogen peroxide in vivo) and glutathione reductase which have been shown to correlate with a predisposition to liver disease10,11. The authors speculate that a systematic analysis of the components of their model could predict other areas where variation could alter the system in ways that render it more susceptible to insult and hence be the basis for idiosyncratic DILI.


Figure 1 Overview of DILI-sim A. Schematic overview of the key biological processes represented in DILI-sim B. Overview of different modules within DILI-sim. Each module is itself a model that captures a specific area of relevant biology, pharmacology and metabolism. This modular approach to DILI-sim allows the overall model to be built in manageable, testable pieces C. Knowledge management aspects of DILI-sim. Under each of the models, the supporting evidence is explicitly captured and hence the model acts as a highly structured knowledge repository
Although this model contains a set of 112 coupled equations, the biochemistry it describes is limited with respect to the breadth of biology and pharmacology which are thought to potentially contribute to idiosyncratic DILI. More recently, a new collaborative initiative called ‘DILI-sim’ has been started with the ambition to develop a more complete model for the prediction of DILI (Paul Watkins, personal communication). DILI-sim was started through a Cooperative Research and Development Agreement (CRADA) between the Food and Drug Administration (FDA), the systems modelling company Entelos (www.entelos.com) and the The Hamner Institutes for Health Sciences but has recently attracted additional industrial and academic collaborators. The scope of DILI-sim is large, encompassing a multi-compartment model of the liver, drug metabolism pathways and immune com – ponents (Figure 1a). DILI-sim is really a ‘meta-model’ as each process represented within it is itself a model (Figure 1b). Using DILI-sim it will be possible to simulate the responses of a whole host of ‘virtual patients’ and in doing so consider how a range of risk factors could affect the response of the liver to a pharmaceutical. Moreover, DILI-sim can also represent rat and mouse species and strains allowing experimental validation of the model as well as facilitating the translation of preclinical data to patients.
DILI-sim is not the only collaborative effort using systems biology in the area of liver biology. The Virtual Liver Network (VLN) is a major German national initiative, funded by the BMBF (Bundesministerium für Bildung und Forschung) to the tune of EUR 43 million over five years. The network brings together over 70 groups and some 50 Principal Investigators distributed across Germany through a number of activities described in nine separate but interconnected work packages (Adriano Henney, personal communication). Although not specifically focused on DILI, the VLN will, like DILI-sim, also attempt to build an organ-scale model of the liver which of course may be applied to the problem of predicting and understanding idiosyncratic DILI.
Currently idiosyncratic DILI is unpredictable, but this doesn’t mean that it is inherently so. The multi-factorial nature of idiosyncratic DILI means that a reductionist approach to its prediction is almost bound to fail. Developing systems models for the prediction of idiosyncratic DILI is a promising new development precisely because such approaches can integrate the multitude of factors which we know contribute to idiosyncratic DILI. The usual and the unusual can be considered and hence idiosyncratic response predicted together with the circumstances under which they could occur and so, unlike a real clinical trial, a virtual one can in some sense consider everyone! This article has highlighted a couple of new initiatives that are using dynamic simulations to tackle the problem of predicting idiosyncratc DILI. Such models will by necessity be large, complex and costly to develop and hence may be best tackled in collaborative efforts like DILI-sim and the VLN; however, there is a more compelling reason for collaboration to be at the heart these initiatives. The complexity of models reflects the state of knowledge within the area and therefore all of the individual elements, relationships and assumptions in something like DILI-sim can be traced back to an original source (Figure 1c, page 62). Hence these models are not just simulations but also ‘knowledge repositories’ and may provide a ‘neutral’ platform of evidence for future debates around idiosyncratic DILI. Ultimately, the first generations of models for the prediction of idiosyncratic DILI may fail, but in doing so will highlight the gaps in understanding that become the foundation of new investigative research.
Acknowledgements
The author would like to thank Paul Watkins for many discussions on DILI-sim and Scott Siler for the provision of Figure 1. The author also thanks Adriano Henney for discussions about the VLN as well as Howard Mellor and Stephanie Roberts for critical reading and help in putting this manuscript together.
References
1. Chalasani N and Björnsson E (2010). Risk Factors for Idiosyncratic Drug-Induced Liver Injury. Gastroenternology 138, 2246-2259
2. Björnsson E and Olsson R (2005). Outcome and prognostic markers in severe druginduced liver disease. Hepatology 42, 481-489
3. Kola I and Landis J (2004). Can the pharmaceutical industry reduce attrition rates? Nature Reviews Drug Discovery 3, 711-716
4. Uetrecht J (2007). Idiosyncratic drug reactions: current understanding. Annu Rev Pharmacol Toxicol 47, 513-519
5. Uetrecht JP (1999). New concepts in immunology relevant to idiosyncratic drug reactions: the ‘danger hypothesis’ and innate immune system. Chem Res Toxicol 12, 387-395
6. Butcher EC (2005). Can cell systems biology rescue drug discovery? Nature Reviews Drug Discovery 4, 461-467
7. Noble D (2008) Computational Models of the Heart and Their Use in Assessing the Actions of Drugs. Journal of Pharmacological Sciences. 107, 107 – 117.
8. Shoda L et al. (2010). The Type 1 Diabetes PhysioLab Platform: a validated physiologically based mathematical model of pathogenesis in the non-obese diabetic mouse. Clinical and Experimental Immunology. May E-Publication ahead of print.
9. Gavaghan D, Coveney PV and Kohl P (Editors) (2009) The virtual physiological human: tools and applications Philos.Transact A. 367
10. Subramanian et al (2008). A systems biology based integrative framework to enhance the predictivity of in vitro methods for drug-induced liver injury. Expert Opin. Drug Saf. 7(6), 647-662
11. Huang YS et al. (2007) Genetic polymorphisms of manganese superoxide dismutase, NAD(P)H:quinione oxidoreductase, glutathione S-transferase M1 and T1, and the susceptibility to drug-induced liver injury. J. Hepatol. 47(1), 128-134
About the Author
Dr David Cook
Dr Cook has a background in biochemistry and molecular biology with a PhD from Imperial College, London and three years of post-doctoral experience. Dr Cook has worked in the pharmaceutical industry for AstraZeneca for more than 13 years. He initially worked in the respiratory and inflammation area and was responsible for identifying the mode-of-action for a new class of immuno – suppressive. He was one of the founders of AstraZeneca’s systems biology group and led the pathway analysis capability in this department for five years. Dr Cook moved into safety sciences four years ago to lead the development of computational biology approaches in support of predictive toxicology and problem-solving.





