The role of chemokines in type 1 diabetes: as assessed by RT-PCR
Posted: 19 March 2008 | Lut Overbergh, Senior postdoctoral fellow, Laboratory of Experimental Medicine and Endocrinology (LEGENDO), at the Catholic University of Leuven and Co-author Prof. Chantal Mathieu M.D. P.h.D., Catholic University of Leuven | No comments yet
Type 1 diabetes is an autoimmune disease, characterised by immune infiltration into the islets of Langerhans, resulting in the destruction of insulin producing b-cells. Over recent years, evidence has been collected on the important role of chemokines in the recruitment of immune cells leading to the pathology of this disease. In this review we discuss the findings on the role of chemokines, as obtained from animal studies. We will focus on the quantification of chemokines and chemokine receptors, making use of the innovative real-time quantitative PCR technique.
Type 1 diabetes is an autoimmune disease, characterised by immune infiltration into the islets of Langerhans, resulting in the destruction of insulin producing b-cells. Over recent years, evidence has been collected on the important role of chemokines in the recruitment of immune cells leading to the pathology of this disease. In this review we discuss the findings on the role of chemokines, as obtained from animal studies. We will focus on the quantification of chemokines and chemokine receptors, making use of the innovative real-time quantitative PCR technique.
Type 1 diabetes is an autoimmune disease, characterised by immune infiltration into the islets of Langerhans, resulting in the destruction of insulin producing b-cells. Over recent years, evidence has been collected on the important role of chemokines in the recruitment of immune cells leading to the pathology of this disease. In this review we discuss the findings on the role of chemokines, as obtained from animal studies. We will focus on the quantification of chemokines and chemokine receptors, making use of the innovative real-time quantitative PCR technique.
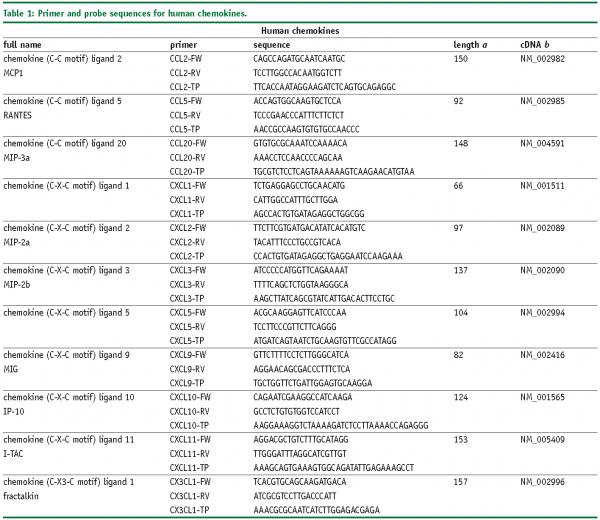
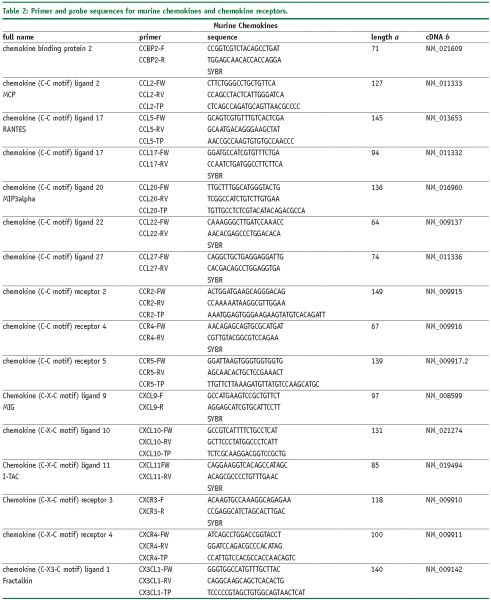
Abbreviations
FW – Forward primer
RV – Reverse primer
TP – TaqMan® probe
SYBR, SybrGreen™ assay.
a – Amplicon length in base pairs
b – Genbank accession number of cDNA and corresponding gene, more information is available at www.ncbi.nlm.nih.gov
Real-time PCR: an introduction
The real-time PCR (RT-PCR) technique is an especially sensitive and accurate technique for the quantification of mRNA levels in cells, tissues and tissue biopsies. Since its invention in the early nineties,1,2 its use has increased exponentially and it is now the method of choice for quantification of mRNA levels. For the quantification of chemokine and chemokine receptor levels specifically, it is ‘the golden standard’, as these factors are often expressed at very low levels in cells or tissues of interest. Currently different DNA polymerases and a wide range of different detection chemistries are available, the original method developed for the quantification of mRNA made use of the intrinsic capacity of the Thermus Aquaticus (Taq) polymerase to amplify DNA and simultaneously, the 5’ nuclease activity of this enzyme to cut an internally annealing dual-labelled fluorescent probe. This degradation of the dual-labelled probe gave rise to a fluorescent emission, which could be measured during the course of the PCR reaction1,2,3 this is the ‘so-called’ specific detection method. A second widely used method makes use of non-specific detection chemistry, such as SYBR Green. In this case, all dsDNA amplified during the reaction is detected.
Therefore, it is very important to include additional controls to distinguish between the target sequence of interest on one hand, a-specific products or primer dimers on the other4.
In both applications, PCR amplification and data acquisition (by measuring the increase in fluorescent emission as the PCR reaction proceeds) is performed in one single step, since fluorescent emission is measured continuously during the PCR reaction, in real-time. In case of non-specific chemistries, a post-PCR melting curve analysis needs to be performed. Quantification of the target of interest is based the increase in fluorescent emission as a direct measure for the amount of amplified PCR product5.
Different factors have lead to the widespread use of real-time PCR assays, being present as a routine technique on almost every laboratory bench. First of all, as opposed to the classical PCR which only resulted in ‘qualitative’ data, it is a ‘quantitative’ method, giving a very good estimation of initial mRNA copy numbers. Secondly, PCR amplification and detection are performed in a single step, resulting in short analysis time and high throughput. Thirdly, quantification is possible over a wide dynamic range. Fourthly, reproducibility and sensitivity of the method are very high, making the method suitable for analysis on very small amounts of cells or tissue samples. This enhanced sensitivity is of particular interest for research and diagnosis in the field of immunology, where cytokines and chemokines are often expressed at very low levels and only small amounts of sample material can be used6,7. Finally, its widespread use has been facilitated by the introduction of second generation instrumentation as well as the design of alternative chemistries; with the natural competition between different commercial suppliers making the technique affordable for almost every scientific and clinical laboratory.
Studies in animal models
Many studies in animal models, most of them performed in the non-obese diabetic (NOD) mouse, have aimed at gaining a better insight into the role of chemokines in the initiation or development of type 1 diabetes8. Early studies in mice have already shown an important role for chemokines in the recruitment of effector T-cells9,10,11 and regulatory T-cells12 to the islets of Langerhans. Furthermore, the CC chemokine gene family is located in the Idd4 diabetes susceptibility locus13, providing genetic evidence for the involvement of chemokines in the pathology of the disease. Based on these findings it is very likely that different chemokines are associated with T-cell differentiation and recruitment as well as with diabetes susceptibility in NOD mice.
The site of immune attack, namely the islets of Langerhans, has been the subject of many studies investigating the expression profile of chemokines, performed in different mouse models.
A first study identified the expression of a whole range of chemokines in islets from NOD-SCID mice, including CXCL10 (IP10), CCL22 (MDC; macrophage-derived chemoattractant), CCL21 (SLC; secondary lymphoid tissue chemokine), CCL3 (MIP1a), CCL17 (TARC; thymus and activation regulated chemokine) and CCL2 (MCP1)14. In addition, high levels of CCL2 (MCP1), CXCL10 (IP10), CCL20 (MIP3a) and the cytokine IL15 were measured in pancreatic islets from pre-diabetic NOD mice, as well as in islet isografts after transplantation into diabetic NOD mice15,16. Another interesting model is the RIP-GP mouse model, a transgenic mouse expressing the glycoprotein of the lymphocytic choriomeningitis virus (LCMV) under the control of the rat insulin promoter17. These mice develop severe insulitis upon infection with LCMV. Different chemokines, namely CXCL10 (IP10), CXCL9 (MIG), CCL2 (MCP1) and CCL5 (RANTES), were highly induced in the pancreatic islets of these mice, concurrent with insulitis development. Also, the chemokine receptors CCR5, CCR2 and CXCR3 were detected specifically in the islets of these transgenic mice, but not in control islets13,18. Interestingly, most of these studies again made use of the real-time PCR technique to measure chemokine transcript levels.
Neutralisation of CXCL10 (IP10) suppresses the development of diabetes in NOD mice, at least after cyclophosphamide induced disease acceleration in NOD mice, through enhanced b-cell proliferation in insulitis19, indicating an important role for this chemokine in the development of diabetes. In contrast, when expressed outside of the pancreas, IP10 can suppress diabetes. This was shown by Christen et al., both in NOD and in a transgenic RIP-LCMV model of autoimmune diabetes. A selective expression of IP-10 was seen in the pancreatic draining lymph node20. In both cases, infection with the LCMV virus whilst the auto aggressive process is already in progress was able to arrest type 1 diabetes progression. This study highlights the importance of correct location, time and expression levels20. Transgenic mice with b-cell specific expression of IP10 did not exhibit spontaneous diabetes when crossed with the RIP-LCMV mice. However, the onset of virus induced diabetes was much earlier in these mice, providing evidence for the involvement of chemokines in the attraction of lymphocytes to the inflamed target site21. Moreover, a direct role for IP10 was convincingly shown in CXCR3 knockout mice, which showed a delayed diabetes onset and prolonged islet graft survival. A similar finding was observed making use of alpha-rIPa (anti-IP10) mAb therapy18,22. In addition, an association between increased IP10 and MCP1 levels was consistent, with a decrease in apoptosis and preserved glucose stimulated insulin release in IRF1 deficient mice23.
An important role in islet infiltration is also evident for CCL2 (MCP1), as shown by different studies24. It has been suggested that MCP1 plays a role in the initiation of type 1 diabetes, contributing to early islet infiltration by attracting lymphocytes. This was shown in the NOD mouse model25. In transgenic mice over-expressing MCP1 and under the control of a rat insulin promoter, a direct role for MCP1 in pancreatic monocytic infiltration was shown26.
CCL3 (MIP1a) is another chemokine shown to play a role in type 1 diabetes development. In NOD mice deficient in MIP1a (NOD.MIP1a knockout), diabetes incidence was significantly reduced and delayed. In addition, the ratio of MIP1a /MIP1‚ in the pancreas appeared to be important during the initial stages of islet mononuclear cell infiltration in determining the nature of insulitis progression in NOD mice25.
Concluding from these studies, chemokines clearly plays a major role in b-cell destruction. The advances recently obtained in this field have been greatly aided by the development of real-time RT-PCR assays for the detection of chemokines in the islets of Langerhans. Indeed, this technique made it possible to investigate and quantify a multitude of chemokines expressed in the islets. Consequently, this has aided to our understanding of the pathology of type 1 diabetes. Combined with numerous studies, deleting or over expressing one chemokine by use of knockout and transgenic mice, or by use of systemic administration of chemokines, or their specific antibodies, it is clear that the expressed chemokines, indeed play an important role in the development of type 1 diabetes. Most probably, however, it will be the cooperation of many cytokines and chemokines which is crucial in mediating the initiation and development of insulitis and diabetes outcome – in mice as well as in humans.
Conclusions
The invention of real-time PCR has not only enormously simplified the quantification of DNA and RNA, it has also resulted in much more accurate and reliable data. This had a great impact in the field of molecular research and diagnostics, since enormous amounts of data could be obtained within a very short research time. In fundamental research on type 1 diabetes, the technique has significantly improved our understanding of b-cell destruction, for instance by the high throughput analysis of chemokine expression levels at the site of immune attack, ie. the islets of Langerhans. The decreased costs for thermal cycling instruments, as well as for the necessary reagents to perform the reactions, have obviously assisted in its rapid, not to say exponential, increase in use. It is beyond any doubt that this technique has become the ‘golden standard’ for the detection and quantification of DNA and RNA in fundamental research, pre-clinical and clinical settings; including the quantification of chemokines.
Although real-time PCR assays by themselves are characterised by high precision and reproducibility, the accuracy of the data obtained relies largely on several other factors. Indeed, it is not sufficient to simply extend one’s knowledge of classic endpoint PCR. Many other controls have to be included to obtain accurate results. Issues to be taken into account for instance are sample preparation, quality of the RNA, choice of a relevant housekeeping gene, correct normalisation of the samples, experimental setup and accurate data analysis5,27.
Most probably, since RT-PCR has become a routine technique present in almost every laboratory, its use is expected to increase even more during the coming decade. However, it remains crucial that specific controls as to the validity of each specific assay are taken into account. Apart from this, as we have reached the post-genomic area, one has to realise that when making use of this technique, no conclusions can be made to the functional implications at the protein level. Combining RT-PCR results with proteomic results, will certainly unearth a more complete view of the dynamic alterations taking place in a cell or tissue of interest under certain physiological or pathological conditions.
It is to be expected that in the field of proteomics analysis, with the major advances being made during the last decade on highly sophisticated mass spectrometry instrumentation, the analysis of translational and post-translational control (eg. post-translational modifications, proteolytic cleavage) will evolve rapidly and enlarge our view on the function and importance of chemokines in type 1 diabetes. These, in combination with RNA based techniques as discussed herein, will greatly aid in filling the gaps that still remain in our understanding of this complex disease.


References
- Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 6(10), 986-94 (1996).
- Higuchi R, Dollinger G, Walsh PS and Griffith R. Simultaneous amplification and detection of specific DNA sequences. Biotechnology 10, 413-417 (1992).
- Holland PM, Abramson RD, Watson R, Gelfand DH. Detection of specific polymerase chain reaction product by utilizing the 5’3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci U S A. 88(16), 7276-80 (1991).
- Wittwer CT, Herrmann MG, Moss AA, Rasmussen RP. Continuous fluorescence monitoring of rapid cycle DNA amplification. Biotechniques 22, 130-131, 134-138 (1997).
- Overbergh L, Giulietti A, Valckx D, Decallonne R, Bouillon R, Mathieu C. The use of real-time reverse transcriptase PCR for the quantification of cytokine gene expression. J Biomol Tech. 14(1), 33-43 (2003).
- Silva TA, Garlet GP, Lara VS, Martins W Jr, Silva JS, Cunha FQ. Differential expression of chemokines and chemokine receptors in inflammatory periapical diseases. Oral Microbiol Immunol 20, 310-316 (2005).
- Carvalho-Gaspar M, Billing JS, Spriewald BM, Wood KJ. Chemokine gene expression during allograft rejection: Comparison of two quantitative PCR techniques. J Immunol Methods 301, 41-52 (2005).
- Atkinson MA and Leiter EH. The NOD mouse model of type 1 diabetes mellitus: As good as it gets? Nat Med. 5, 601-604 (1999).
- Bradley LM, Asensio VC, Schioetz LK, Harbertson J, Krahl T, Patstone G, Woolf N, Campbell IL, Sarvetnick N. Islet-specific Th1, but not Th2, cells secrete multiple chemokines and promote rapid induction of autoimmune diabetes. J Immunol. 162(5), 2511-20 (1999).
- Kim SH, Cleary MM, Fox HS, Chantry D, Sarvetnick N. CCR4-bearing T cells participate in autoimmune diabetes. J Clin Invest. 110(11), 1675-86 (2002).
- Savinov AY, Wong FS, Stonebraker AC, Chervonsky AV. Presentation of antigen by endothelial cells and chemoattraction are required for homing of insulin-specific CD8+ T cells. J Exp Med. 197(5), 643-56 (2003).
- Szanya V, Ermann J, Taylor C, Holness C, Fathman CG. The subpopulation of CD4+CD25+ splenocytes that delays adoptive transfer of diabetes expresses L-selectin and high levels of CCR7. J Immunol. 169(5), 2461-5 (2002).
- Grattan M, Mi QS, Meagher C, Delovitch TL. Congenic mapping of the diabetogenic locus Idd4 to a 5.2-cM region of chromosome 11 in NOD mice: identification of two potential candidate subloci. Diabetes. 51(1), 215-23 (2002).
- Giarratana N, Penna G, Amuchastegui S, Mariani R, Daniel KC, Adorini L. A vitamin D analog down-regulates proinflammatory chemokine production by pancreatic islets inhibiting T cell recruitment and type 1 diabetes development. J Immunol. 173(4), 2280-7 (2004).
- Chen MC, Proost P, Gysemans C, Mathieu C, Eizirik DL. Monocyte chemoattractant protein-1 is expressed in pancreatic islets from prediabetic NOD mice and in interleukin-1 beta-exposed human and rat islet cells. Diabetologia. 44(3), 325-32 (2001).
- Cardozo AK, Proost P, Gysemans C, Chen MC, Mathieu C, Eizirik DL. IL-1beta and IFN-gamma induce the expression of diverse chemokines and IL-15 in human and rat pancreatic islet cells, and in islets from pre-diabetic NOD mice. Diabetologia. 46(2), 255-66 (2003).
- Ohashi PS, Oehen S, Buerki K, Pircher H, Ohashi CT, Odermatt B, Malissen B, Zinkernagel RM, Hengartner H. Ablation of “tolerance” and induction of diabetes by virus infection in viral antigen transgenic mice. Cell. 65(2), 305-17 (1991).
- Frigerio S, Junt T, Lu B, Gerard C, Zumsteg U, Holländer GA, Piali L. Beta cells are responsible for CXCR3-mediated T-cell infiltration in insulitis. Nat Med. 8(12), 1414-20 (2002).
- Morimoto J, Yoneyama H, Shimada A, Shigihara T, Yamada S, Oikawa Y, Matsushima K, Saruta T, Narumi S. CXC chemokine ligand 10 neutralization suppresses the occurrence of diabetes in nonobese diabetic mice through enhanced beta cell proliferation without affecting insulitis. J Immunol. 173(11), 7017-24 (2004).
- Christen U, Von Herrath MG. IP-10 and type 1 diabetes: a question of time and location. Autoimmunity. 37(5), 273-82 (2004).
- Rhode A, Pauza ME, Barral AM, Rodrigo E, Oldstone MB, von Herrath MG, Christen U. Islet-Specific Expression of CXCL10 Causes Spontaneous Islet Infiltration and Accelerates Diabetes Development. J Immunol. 175(6), 3516-24 (2005).
- Baker MS, Chen X, Rotramel AR, Nelson JJ, Lu B, Gerard C, Kanwar Y, Kaufman DB.Genetic deletion of chemokine receptor CXCR3 or antibody blockade of its ligand IP-10 modulates posttransplantation graft-site lymphocytic infiltrates and prolongs functional graft survival in pancreatic islet allograft recipients. Surgery. 134(2), 126-33 (2003).
- Baker MS, Chen X, Rotramel AR, Nelson JJ, Kaufman DB. Interferon regulatory factor-1 down-regulates cytokine-induced IP-10 expression in pancreatic islets. Surgery. 34(2), 134-41 (2003).
- Schroppel B, Zhang N, Chen P, Chen D, Bromberg JS, Murphy B. Role of donor-derived monocyte chemoattractant protein-1 in murine islet transplantation. J Am Soc Nephrol. 16(2), 444-51 (2005).
- Cameron MJ, Arreaza GA, Grattan M, Meagher C, Sharif S, Burdick MD, Strieter RM, Cook DN, Delovitch TL. Differential expression of CC chemokines and the CCR5 receptor in the pancreas is associated with progression to type I diabetes. J Immunol. 165(2), 1102-10 (2000).
- Grewal IS, Rutledge BJ, Fiorillo JA, Gu L, Gladue RP, Flavell RA, Rollins BJ. Transgenic monocyte chemoattractant protein-1 (MCP-1) in pancreatic islets produces monocyte-rich insulitis without diabetes: abrogation by a second transgene expressing systemic MCP-1. J Immunol. 159(1), 401-8 (1997).
- Giulietti A, Overbergh L, Valckx D, Decallonne B, Bouillon R and Mathieu C. An overview of Real-Time Quantitative PCR: Applications to Quantify Cytokine Gene Expression. Review. Methods 25, 386-401 (2001).





