Human Embryonic Stem Cells: Progress in culture and maintenance
Posted: 29 September 2008 | Fergus R. McKenzie, Programme Manager, ITI Life Sciences | No comments yet
The pharmaceutical industry is at a crossroads: Research and Development costs continue to rise, whilst the pipeline of ‘blockbuster’ drugs is looking decidedly sparse for many of the major pharmaceutical companies. This state of affairs is compounded by the high attrition rates (>80%) of drug candidates, when they get to clinical trials. Thus, the major pharmaceutical companies are keen to apply new technologies that allow faster decision making (proceed with development of a drug candidate, or stop at an earlier point) and ultimately increase the number of new medicines that reach the marketplace.
The pharmaceutical industry is at a crossroads: Research and Development costs continue to rise, whilst the pipeline of ‘blockbuster’ drugs is looking decidedly sparse for many of the major pharmaceutical companies. This state of affairs is compounded by the high attrition rates (>80%) of drug candidates, when they get to clinical trials. Thus, the major pharmaceutical companies are keen to apply new technologies that allow faster decision making (proceed with development of a drug candidate, or stop at an earlier point) and ultimately increase the number of new medicines that reach the marketplace.
The pharmaceutical industry is at a crossroads: Research and Development costs continue to rise, whilst the pipeline of ‘blockbuster’ drugs is looking decidedly sparse for many of the major pharmaceutical companies. This state of affairs is compounded by the high attrition rates (>80%) of drug candidates, when they get to clinical trials. Thus, the major pharmaceutical companies are keen to apply new technologies that allow faster decision making (proceed with development of a drug candidate, or stop at an earlier point) and ultimately increase the number of new medicines that reach the marketplace.
Most, if not all pharmaceutical companies are committed to cell-based high throughput screening (HTS). This is because a functional read-out in a more physiologically relevant context is likely to provide information that better predicts the chances of success of lead compounds. Indeed following this thought process, cell based screening is now used in most areas of drug discovery including target validation, primary screening, secondary screening (hit to lead) and ADMET (adsorption, distribution, metabolism, excretion and toxicity) studies1,2. This article provides an overview of the role of hES cells in the drug discovery process. Readers are directed to recent reviews on the subject for further details17,18.
Cell-Based Screening: What Cell Types?
Pharmaceutical companies have a range of criteria that have to be met in order to integrate cells or cell-based assays into their drug screening regime. A cell ‘wish list’, in no particular order, may look like the following:
- Physiologically relevant, therefore human and appropriately differentiated
- Available in large quantities, hence amenable to automated production
- Genetically stable and capable of supporting genetic manipulation (eg ectopic expression of a drug target)
- Capable of supporting a read-out assay
- Have an acceptable cost/benefit
- Ethically acceptable source
Arguably the most preferred cell types to pharmaceutical companies are: human hepatocytes, cardiomyocytes, neuronal cells and insulin-producing pancreatic cells. However, current supply of such cell types (e.g. from cadavers and biopsies) will never be able to match demand. Hence a source of such cells is highly desired. A further advantage of having a ready supply of specific human cell types is that such cells may be used in the field of regenerative medicine, where patients receive cells in an attempt to reconstruct organs and tissue that have atrophied or been destroyed as a result of degenerative or other illnesses or trauma.
Embryonic stem cells: The solution?
Embryonic stem cells are capable of unlimited self-renewal (immortal), can be instructed to differentiate into all somatic cell types, are amenable to genetic modification and therefore present themselves as a solution to the physiologically relevant human cell supply problem. The initial isolation of mouse embryonic stem cells occurred in 19813,4. Seventeen years later, human embryonic stem cells were generated, following a similar derivation protocol5,6. Those out with the field suspected that the research on mouse ES cells would be readily applicable to human ES cells and soon, scientists would be generating large numbers of hES cells for basic research, the drug discovery industry, and, of course, regenerative medicine.
Unfortunately, this proved not to be the case. hES cells are significantly different to mouse ES cells in their population doubling times, growth conditions and response to known differentiation factors. As a consequence, until recently, hES cells have remained in the domain of a limited number of specialist laboratories and have not crossed over to the realm of either generalist laboratories or big pharma. A summary of the advantages and disadvantages of hES and their derivatives is presented in Table 1.
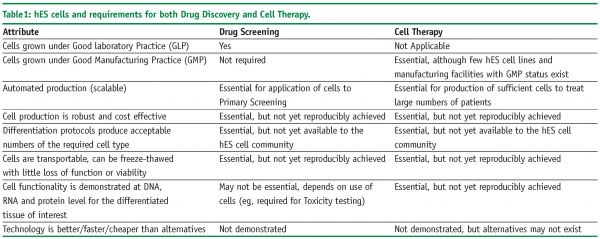

Unlike most other cell types and tissue sources, hES cells have the potential to be employed in the drug discovery industry, as well as being strongly linked to the production of new therapeutic regimes for regenerative medicine. Indeed, many of the requirements for cell therapy represent extensions in quality to what is required for general use in drug discovery. However, this potential will only be realised if the growth and maintenance of hES cells can be rendered robust, with simplified cell media and automated production methods in place. These will be explored below.
Progress in hES cell culture: Media
In common with mouse ES cells, human ES cells were derived on a supporting feeder layer of mitotically inactivated (unable to further increase in number) mouse fibroblasts. This feeder layer of mitotically inactive cells is thought to provide several functions:
- Provides an anchoring point for cell attachment
- Provides factors through either secretion into the cell culture media or through direct cell-cell contact that promote cell growth and division
- Provides factors through either secretion into the cell culture media, or through direct cell-cell contact that prevent hES cell differentiation
In some instances, it is possible to grow hES cells in the absence of the fibroblast feeder layer, but with the addition of media that has been ‘conditioned’ by the feeder layer, such that factors secreted by the feeder layer are present and able to regulate hES cell growth. However, they still require physical contact with cell surface components (extracellular matrix, ECM) which have to be coated onto the tissue culture surface before the cells are added.
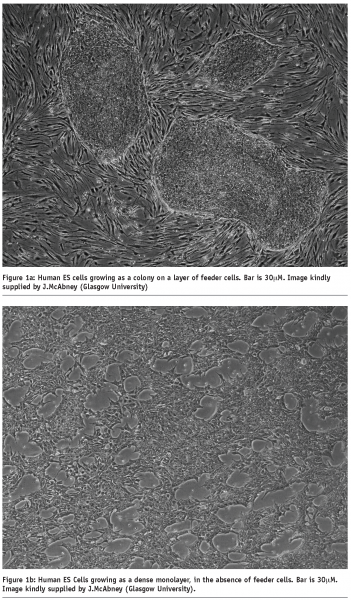

In part a (of Figure 1), we see three separate colonies of undifferentiated hES cells, growing on top of a layer of feeder cells. In part b (of Figure 1), the hES cells have been removed from the feeder layer and placed in cell culture conditions that allow them to attach to the ECM-coated plastic dish as a monolayer, and remain in an undifferentiated state.
In addition, hES cells are cultivated in the presence of expensive growth factors and 10-15% serum. However, these cell culture conditions are not amenable to drug discovery, since the presence of growth factors, serum and ECM or feeder cells would all confound the ability to make predictive measurements corresponding to the hES cells or hES cell derivatives. We have thus seen a move towards the stepwise replacement/ improvement of undesirable elements in hES cell culture protocols.
The final solution that many companies are seeking is the ability to grow hES cells in the absence of feeder layer, in the presence of a fully defined cell culture media that completely lacked serum or other undefined components. Furthermore, the ability to grow the cells continuously and subculture into appropriate cell culture dishes is essential.
At the moment, several products exist on the market which represents significant steps forward in the standardisation of hES cell culture:
a. STEMPRO® – sold by Invitrogen and is a fully defined, serum and feeder-free medium which allows expansion of hES cells
b. mTeSRTM1 – sold by StemCell Technologies and is a serum-free, fully defined (although it does include animal-sourced proteins) medium that does not require feeder cells
In both of the above commercial media, the challenge of hES cell attachment to the plastic dish is solved by the inclusion of an ECM that provides a ‘bridge’ between the culture dish and the hES cells. Indeed, this is a key issue relating to both the cell culture media and the ability to automate hES cell production. hES cells tend to grow in colonies or clumps. If we use enzymes to break the clumps apart, we can produce single cell suspensions of hES cells. However, if these cells are plated onto normal tissue culture plasticware, they will enter into programmed cell death (apoptosis), leaving very few attached cells in the dish. A key step forward, was the finding that the inclusion of several inhibitors of intracellular kinases could attenuate the apoptosis of hES cells following cell clump disaggregation7. Both of these media represent a significant advance in the ability to grow hES cells. However, these media may not be applicable to all hES cells, as will be discussed later.
Progress in hES cell culture: Automation
Automation of the drug discovery process was one of the earliest innovations that permitted high throughput screening to become a reality and allow pharmaceutical companies to screen large numbers of compounds for functional effects, within a realistic time frame. However, automation of cell production has lagged behind for a number of reasons; mammalian cells routinely require visual inspection in order to seed cells at an appropriate cell density, cell culture media and plastic-ware is subject to variation and enzymatic digestion is a critical step to which most cells have a sensitive dependence. That said, at least two robotic systems have been employed for the automated production of hES cells:
a. The Hamilton Microlab Cellhost system8
b. The Automation Partnerships’ SelecT system9
The Cellhost system (see Figure 2a), produced by Hamilton, is based on their Microlab STAR workstation that has been adapted to cell culture. The essence of the system is a plate-lifter that tilts cell culture plates and allows the complete removal of supernatant from the cells. The CellHost has been used to culture mouse ES cells continuously for four weeks8. The Compact SelecT (see Figure 2b) from the Automation Partnership is the little brother of the SelecT and was designed with mammalian cell culture in mind. This system used a robotic anthropomorphic arm to mimic human movement and similarly to the Hamilton system, has been used to cultivate hES cells for multiple passages, retaining hES cell quality9.


A variety of additional automation systems exist10, such as the Cell-IQ system (Chip-Man Technologies Ltd, Tampere, Finland) and the BioCel 1800 system (www.velocity11.com) in place at the Institute for Stem Cell Therapy and Exploration of Monogeneic Diseases (www.istem.eu), in addition to scale-up methods with bioreactors11 and culture vessels designed to provide a substrate in three dimensions12. Indeed it is highly possible that it will prove difficult to differentiate hES cells all the way to terminally differentiated complex cell types without a move towards more three dimensional culture techniques12.
All hES cells have not been created equal
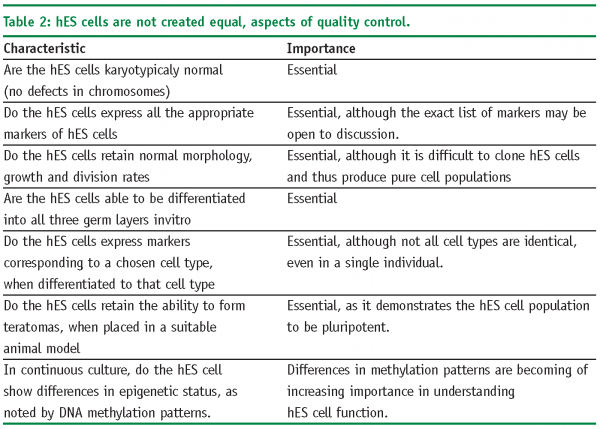
World-wide estimates of hES cell derivation suggest that there are approximately 200 hES cell lines (Estimate made in 200413). The figure in 2008 may now be approaching 300. Although all hES share a similar morphology, expression of transcription factors such as Oct3/4 which indicate pluripotency and have the ability to differentiate into all three germ layers (endo-, ecto- and mesoderm) in-vitro, the differentiation potential of each cell line can vary significantly. This is borne out by data relating to population doubling times for hES cells in culture which vary in the literature from 24h to as much as 48h14. Further evidence of hES cell line differences come from the scientific literature, which is beleaguered by an inability of scientists to repeat their colleagues published work on a range of different hES cell lines (informal survey taken at scientific meetings during 2006-8, Pers Comm).
As a consequence of the variation seen between hES cell populations, the cell culture requirements for each hES cell line can also vary. To what can we attribute this variation? Firstly, although following similar protocols, hES cells may represent the environment in which they were derived: Condition and age of the blastocyst starting material, the exact medium employed, the adaptation of the hES cells to the media and the time taken to actually derive an hES cells line. It should also be noted that hES cells are not clonally derived from a blastocyst and hES cell lines should be thought of as a heterogeneous population of cells.
Several initiatives have been undertaken to explore and compare the existing hES cells lines, including the International Stem Cell Initiative (ISCI)15, the NIH Stem Cell Unit and the American Type Culture Collection16. These initiatives are on-going and are investigating some of the parameters noted in Table 2 on page 59.

It is clear therefore, that hES cells represent a complex model system for drug discovery. Indeed, the cells are at a stage in their technology development where many questions remain to be adequately addressed.
Whilst these initiatives to compare hES cell lines are to be lauded, it should be noted that many of the studies by associated scientific groups are using their own cell culture conditions, which may make it difficult to draw overall conclusions other than there is significant heterogeneity in the field of hES cells and this is set to continue until a range of standard parameters are accepted. In the case of pharmaceutical companies, who are usually interested in technology development the simplest option may be to chose one set of hES cells, cell culture medium and automated production methods that have been shown to work to acceptable levels. Of course, we have to ask the question of the importance of this level of hES cell characterisation. It could be argued that we would like to have answers to all of the attributes noted in Table 2, for cell therapy. However, if hES cells are going to be used for drug discovery, the question of epigenetic status and DNA methylation (for example), may be unimportant as such cells may still behave in-vitro in a manner that is functionally equivalent to the desired cell type.
hES Cell Differentiation
As hES cells start to move along a differentiation pathway towards a specific cell type, their ability to self-replicate starts to diminish. At the other end of the spectrum from hES cells, a fully differentiated cell type such as a cardiomyocyte has little or no replicative capacity. Hence, production of large numbers of cardiomyocytes will require either a large number of hES cells to start with, or efficient differentiation and expansion processes. Early attempts to differentiate hES cells in-vitro involved the production of ‘embryoid bodies’ which are essentially ‘clumps’ of hES cells which undergo spontaneous differentiation. The cell-cell contact and media conditions employed, initiated a differentiation process in which the hES cells could become a range of different cell types. However, this method of differentiation is difficult to control.
In view of the differences in hES cell lines described above, it is no surprise to note that robust protocols to efficiently and specifically differentiate hES cells into lineages from all three germ layers do not yet exist.
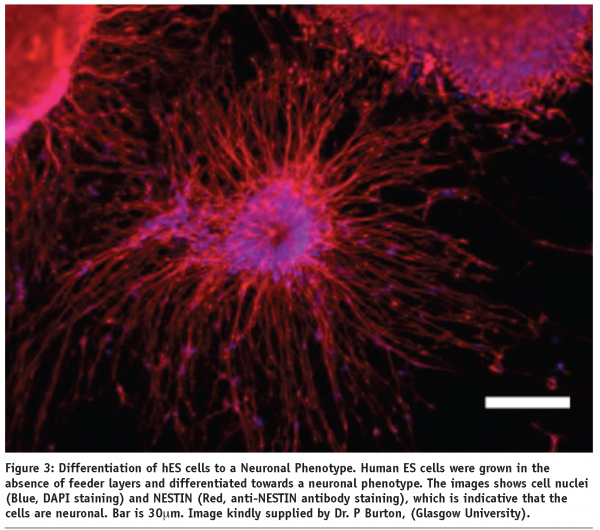

As shown in Figure 3, many laboratories are able to produce a differentiated cell types at a small scale. However, the expression of protein markers indicative of a desired cell type does not mean that the cell has the functional equivalence of that cell type, as measured in-vivo.
This is arguably the least understood area of hES cell control and the rate limiting step for the uptake of hES by the drug screening industry. Despite several reports over the last decade, the translation of differentiation conditions from individual cell culture dishes, where an often minor proportion of the hES population eventually differentiates into a chosen cell type, has proved laborious17. Efforts to produce single cell types should probably concentrate less on continuous cell culture and may favour highly lineage-specific scale up methods such as suspension bioreactors with cells either grown in aggregates, or on microcarriers18.
Additionally, it should be noted that the functional validation of cells such as neurones, hepatocytes and cardiomyocytes is extremely time consuming, and has to stand comparison with the equivalent tissue harvested from a human patient.
Next Steps: Logical progression, how will the technology advance?
The culture conditions for hES cells have been described above. However, recent advances suggest that adult mature somatic cells (e.g. skin-derived fibroblasts) have the ability to be ‘re-programmed’ to de-differentiate and become cells that bear several hallmarks of hES cells (expression of key pluripotency markers, ability to self-renew, capacity to differentiate into the three germ layers)19,20. Many technical barriers have to be over-come, but it will be interesting to see whether the technology trajectory facing these so-called induced pluripotent stem (iPS) cells follows a similar path and timescale to that taken by hES cells.
All of the issues discussed support a need for the development of a technology “roadmap” for hES cell culture. This would aim to identify and recommend best practice for hES cell culture and differentiation. Consequently, we might expect to see an increased rate of improvement in hES cell expertise resulting in many of the factors required for this technology to enter into wide-scale use in the drug-screening industry including:
- Cheaper cost of cell media
- Robust manufacturing processes to deliver high-quality differentiated cell types
- The hES cell culture arena becoming universally accessible, not just in the specialist hES cell laboratories in academia, biotech or (eventually) the large pharmaceutical companies
Conclusion
The last few years have seen significant advances in the culture of hES cells, these are evident in the fields of automated cell production, cell media formulation, removal of feeder cells and improved differentiation protocols. At the moment, hES cell research is in a phase where funding is available from a number of sources. It can only be expected that over a three-five year timescale we will see further stepwise improvements in hES cell husbandry, as well as the increased application of hES cells and their derivatives to the cell-based drug screening industry. Ultimately, we are getting close to the goal of delivering on new medicines and treatments for degenerative illness however, such treatments come directly from hES cells, or from ES-cell like cells, remains to be seen.
Acknowledgements
The author thanks all members of the ITI Life Sciences Stem Cell Technologies (SCT) team for their commitment and dedication to the SCT programme
References
- M. Bickle: Live cell high content screening in drug development. European Pharmaceutical Review, 4:pp. 54-60, 2008.
- Cell/tissue Culture Supplies: Global Strategic Business Report. Global industry Analysts Inc. 2006
- M.J. Ewans and M.H. Kaufman: Establishment in culture of pluripotential cells from mouse embryos. Nature, 292:pp. 154-156, 1981.
- G.R. Martin: Isolation of a pluripotential cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl. Acad. Sci. USA. 78(12). pp: 7634-7638, 1981.
- J.A. Thomson et al : Embryonic stem cell lines derived from human blastocysts. Science, 282 (5391):pp. 1145-1147, 1998.
- L.M. Hoffman and M.K. Carpenter: Characterization and culture of human embryonic stem cells. Nat Biotechnol. 2005 23(6):pp. 699-708, 1998.
- K.Watanabe et al: A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat Biotechnol 25:pp 681-686. 2007.
- S.Terstegge et al: Automated maintenance of Embryonic Stem Cell Cultures. Biotechnol and Bioeng. 96(1):pp.195-201, 2007.
- A. Aspegren et al, pers comm..
- S.Narkilahti et al: Monitoring and analysis of dynamic growth of human embryonic stem cells:comparison of automated instrumentation and conventional culturing methods. Biomechanical Engineering Online. 6(11): 2007.
- S.Niebruegge et al: Cardiomyocyte Production in Mass Suspension Culture:Embryonic Stem Cells As a Source for Great Amounts of Functional Cardiomyocytes. Tissue Engineering. 14(10) pp 1-11. 2008.
- S.Pryzborski. Platform technology for routine three dimensional cell culture. Eur Pharma Rev, 3:pp., 2008.
- S.N.Brimble et al: Karyotypic stability, genotyping, differentiation, feeder-free maintenance, and gene expression sampling in three human embryonic stem cell lines derived prior to August 9, 2001. Stem Cells Dev. 13:pp.585-597. 2004
- A.Allegrucci and L.E.Young. Differences between human embryonic stem cell lines. Hum Reprod Update 13 (2): pp 103-120, 2007.
- P.Andrews et al: The International Stem Cell Initiative:towards benchmarks for human embryonic stem cell research. Nat Biotechnol 23:pp.795-797. 2005
- The International Stem Cell Intitative, www.stemcellforum.org; The NIH Stem Cell Unit, http://stemcells.nih.gov/ research/nihresearch/scunit/ ; the American Type Culture Collection, http://stemcells.atcc.org; All web sites accessed on 8th Sept 2008.
- C.W.Poulton and J.M.Haynes: Embryonic stem cells as a source of models for drug discovery. Nature Reviews Drug Discovery 6. pp 605-616. 2007.
- H.Thomson: Bioprocessing of embryonic stem cells for drug discovery. Trends in Biotechnol. 25 (5): pp. 224-230, 2007.
- K.Takahashi et al: Induction of Pluripotent Stem Cells from Adult Fibroblasts by Defined Factors. Cell 131:pp 1-12. 2007.
- I-H Park et al. : Disease-Specific Induced Pluripotent Stem Cells. Cell, 134: pp 1-10, 2008.