The handling and analysis of large scale high content screening data
Posted: 23 May 2007 | | No comments yet
Data management has become one of the central issues in High Content Screening (HCS) as it has high potential within predictive toxicity assessments. In particular, HCS applying automated microscopy requires a technology and system which is capable of storing and analying vast amounts of image and numeric data. HCS data includes comprehensive information about the bioactive molecules, the targeted genes and images, as well as their extracted data matrices after acquisition. Here we describe a bioinformatics solution HCS LIMS (Laboratory Information Management System) for the management of data from different screening microscopes. Additionally, the data handling approaches used in HCS for image converting, compression and archiving of images are discussed.
Data management has become one of the central issues in High Content Screening (HCS) as it has high potential within predictive toxicity assessments. In particular, HCS applying automated microscopy requires a technology and system which is capable of storing and analying vast amounts of image and numeric data. HCS data includes comprehensive information about the bioactive molecules, the targeted genes and images, as well as their extracted data matrices after acquisition. Here we describe a bioinformatics solution HCS LIMS (Laboratory Information Management System) for the management of data from different screening microscopes. Additionally, the data handling approaches used in HCS for image converting, compression and archiving of images are discussed.
Data management has become one of the central issues in High Content Screening (HCS) as it has high potential within predictive toxicity assessments. In particular, HCS applying automated microscopy requires a technology and system which is capable of storing and analying vast amounts of image and numeric data. HCS data includes comprehensive information about the bioactive molecules, the targeted genes and images, as well as their extracted data matrices after acquisition. Here we describe a bioinformatics solution HCS LIMS (Laboratory Information Management System) for the management of data from different screening microscopes. Additionally, the data handling approaches used in HCS for image converting, compression and archiving of images are discussed.
HCS assays are especially useful in the study of cytotoxicity of compounds, because they allow for multiplexing parameters of relevance for cytotoxicity. By using an appropriate combination of fluorescent reagents, cell based HCS assays can also act as a valuable first step prior to studying the toxicological effects of compounds in animal testing; a process that is much more expensive in terms of time resources. Biological databases are becoming increasingly popular, due in part to the large amount of images that are generated by various cell based HCS assays. Image database management systems differ from traditional database management systems in major ways; the first difference is the data complexity, transaction records are composed of simple data elements, i.e. names and numbers. Images, on the other hand, are large complex arrays of values. To handle such data we developed sophisticated information technologies and HCS LIMS1 for collecting and interpreting the enormous volume of biological imaging data produced in HCS laboratories. HCS LIMS was elaborated to screen RNA interference libraries for genes that have an impact on the biological process under investigation (assay), whilst also being applied to chemical screens for toxicity and modification of cellular processes. High Content siRNA screening performed in our group8 was reviewed recently12 and described in particular the work-flow in HCS and current types of RNAi libraries used in mammalian systems. In the next section we will present components for enabling effective web browsing of these large-scale image libraries from automated microscopes. In particular, we describe the HCS LIMS system, data flow and image converter component, which facilitates the archiving of large screening image data.
Challenge for storage in automated microscopy
Viewing biological samples with an automated confocal microscope yields thin optical sections of each sample; allowing precise reconstruction of the entire cell. The resulting data is often complex and full of subtleties, the data becomes even more complex when imaging multiple proteins through multiple, independent channels, and also has a high processing and storage cost. A 2D protein localization image from automated confocal microscopy can require 4MB (1M pixels recorded in two channels) of storage. A 3D localization that records dynamic information can be 10 GB. Today, many modern automated microscopy systems provide up to 100.000 images per day, producing multiple colours simultaneously at an amazingly high resolution that allows for detailed analysis at sub-cellular levels. HCS can easily generate more than one Terabyte in both primary images and metadata. A screening image data set is defined, usually consisting of two parts; the first is an image data file, provided in either binary or ascii text format, the second is information about the image data set (i.e., metadata). Metadata is data about data. Database systems like HCS LIMS should support a range of standard microscope image formats: TIFF 16 bit, TIFF 8 bit, FLEX Evotec, LSM Zeiss, LEI Leica and TIFF Cellomics.
HCS LIMS
HCS LIMS is an example of a way in which to build a web-based customizable bioinformatics system for the managing and analyzing of all areas of high content screening experiments5,6,10,11. The system tracks the complete screening process starting from production of biological/chemical compounds, and their application in assays evaluates their influence on cells by analyzing images generated during experiments. Consequently, these specific aims are recommended for development of such HCS databases:
- A laboratory information management system to keep track of the information that is acquired during the screening production in multititer plates
- Well defined data interfaces for importing and exporting
- A Plug-in Architecture (PA) to connect other bio applications, instrumentation and link to its data without amending the system code
- Interface to allow external applications such as data mining tools to query and read the stored data, as well as write back results.
- Initiation, design and implementation of a user management system that facilitates user authentication and authorisation
- Initiating a database and web portal to browse and upload all screening results
User friendly web browsers, or integrated database clients ensure access to the screening data at every stage (Figure 2). The plate viewer (Figure 3) is one of the main elements of the HCS LIMS system that represents the real multiwell plates used in the HCS laboratory8. The system combines the image information with each well and includes tools to display it graphically (Figure 4). The plates can be selected by their size according to the well number (96, 384, 1536). Each experiment has associated with it several pieces of information associated with it such as the molecular structure, the target gene information and the actual sequence of the genes. In addition, the database can also accept annotations and phenotypic descriptions of screen data by using a controlled vocabulary to log experiences from experiments as well as links to publications that reference the screen.
The HCS LIMS package consists of several single databases that allow the researcher to conduct and track different assays by simply using web browsers or integrated database clients.
The system is a collection of the following modules: Library Production, Library Checker, Library Browser, Booking System, Equipment Manager, Screen Browser and Screen Publisher as presented in Figure 1. Each module of HCS LIMS is assigned and related to the compound (RNAi), which is the central object in the whole system structure.
Modules of HCS LIMS
To use the HCS LIMS, a user must first upload a library of microtiter plates into the system, a first step in data flow is a library data import module. The HCS LIMS input data are biological or chemical compound libraries in excel format (xls) produced mainly by external or internal supplies.2,7
Library Checker
Each incoming data set must be precisely validated by Library Checker (LC) module before it is imported into Library Browser module. LC is a collection of scripts for the examination of input data sets including chemical or biological compounds. LC verifies the library ID, membership of a compound in a batch, and the compound properties. It also compares the result of verification with two independent libraries and highlights the differences. Each incoming library is also cross-checked against duplicates, false position on plates or errors in sequences. Finally, LC performs a duplicate analysis of all compound properties.
Library Browser
Library Browser (LB) guarantees the identification of library and appropriate position of a specific compound on a plate. The location history of each compound in the screen, run and replicated along with reformatting information is then recorded and reconstructed by LB. Within the GUI the user may select the library, plate set and if desired, compound data derived from specific 96, 384 or 1536-well plate. Once a plate is selected, a second window is opened in a plate viewer that provides an easy navigation function within the plate, assisting with the extracting of comprehensive information from wells regarding particular compounds (Figure 3). After all necessary plates have been entered they can be chosen to set up a screening run. A file is then generated and prepared for download which includes a list of all assay plates comprising one screening run and their molecules per well. This file is utilized by the screening robot software to generate a pipeting design file.
Screen Browser
Compound libraries stored in LB are interactively linked with the next module; ‘Screen Browser’ (SB). Its’ data entry begins with the definition of a project, screen, run and all experimental protocols presented in Figure 5; analysing the definitions of biomaterials used, cell culture conditions, experimental treatments, experimental designs, definition of experimental variables, as well as definition of experimental and biological replicates. The end result is a selection of compound libraries for the screen. The user can easily simulate the project hierarchy (Figure 5) via additional interface which simulates cases that exist in real screening processes. SB facilitates remote entry of all information concerning the screen, where users may create associations of labeled extracts and substances, scanned raw images (Figure 4) from microscope and quantification matrices (files with results after image processing).
Screen Publisher (Phenobank)
Phenobank stores information about compound phenotypes and hits in a standardised way; organising screen data into groups. Phenobank also facilitates typical visualisations of phenotypes like scatter plots, bar plots and tables.
SIB
We developed a microscope image converter called Screening Image Browser (SIB)3; this provided us with a convenient method with which to view digital microscope slides produced via screen direct from image storage. Figure 6 presents the basic idea of image converters working with different image formats: TIFF 16 bit, TIFF 8 bit, FLEX Evotec Technologies, LSM Zeiss, LEI Leica and TIFF Cellomics. Using this driver we can access images during a screening process in microscope format direct from microscope storage in same time extract metadata from microscope scan. The image driver can then display this information on image.
User Management System
In order to avoid unauthorised access in a multi-user environment and to control user access we have developed a user management system for HCS LIMS which provides one username and password combination. The purpose of the groups is to define a set of users with common authorisations over elements of the system; in other words, the subsets of plates, projects, screens and runs that a group of users can view or utilise. The groups allow the assignment and management of authorisations without difficulty, but also provide enough control over access of the different users to the subsets, plates, projects, screens and runs.
Data storage and archiving architecture
There are many considerations while designing data storage and archiving for HCS; the principal considerations are the speed of image data transfer, reliability of data, storage capacity, and cost. Here we present (Figure 7) as an example of automated data flow of the HCS facility at the Max Planck Institute in Dresden, Germany8. Data is collected by automated microscopes and stored briefly on redundant local disk cache. Enough local storage cache exists for the capture process of one genome wide run to continue through any network and transfer interruptions that may occur. Data is transferred from the cache via Gigabit networking to heavily redundant disk arrays attached to a 30 node, 60 processor local cluster based on Sun Grid Engine. Several cluster nodes process the captured data and generate the format that is then passed on for image analysis, as well as a compressed format for quick preview. The original data is compressed, archived and transferred to a SAM-FS based system, then removed to tape using a high capacity tape robot. Data is available for processing via multiple software packages in-house using local resources or via additional cluster power through our collaboration with the Dresden Center for Information Services and High Performance Computing9.
Conclusion
The focus of this article has been the description of the growing importance of terabyte-scale image collections in cell and molecular biology research, and identifying information technology challenges that must be addressed in order to maximize the knowledge gained from these collections. We have described relevant work that demonstrates the practicability of creating tools like HCS LIMS to address these challenges in order to advance automated microscopy from a subjective, descriptive practice; based on visual interpretation, to an objective, systematic science that can provide critical knowledge on the spatial and temporal patterns of biological macromolecules. In the developmental phase we worked closely with biological researchers and microscope engineers at the HCS facility of Max Planck Institute in Dresden8 to develop a flexible and extensive system to meet current and future HCS storage requirements.
Acknowledgements
This project was funded in part by the BMBF/InnoRegio/BioMeT grant ‘Förderkennzeichen 03I4035A’ and the Max Planck Society’s inter-institutional initiatives ‘RNA Interference’ and ‘Chemical Genomics Centre’. Marino Zerial and Ivan Baines, both directors at the MPI-CBG, provided the vision for an academic high-content screening centre, and concepts at a very early stage. Thanks to a highly motivated, skilled and dedicated multi-disciplinary team, a unique infrastructure has been developed and established under the leadership of Eberhard Krausz that allows implementation of world-class high content RNAi assay development and screening projects. In particular, we would like to thank Eugenio Fava, Kerstin Korn, Ina Poser, Jan Wagner, Hannes Grabner and Martin Stoeter for our numerous discussions.
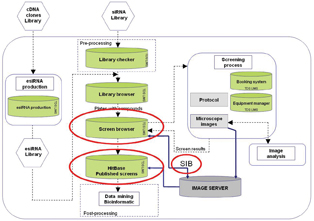

Figure 1: Architecture and modules of the HCS LIMS platform
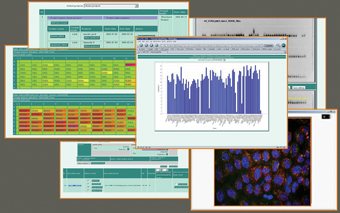

Figure 2: Various visualization procedures such as scatter plots, bar plots or tables simplify assessment of screen results
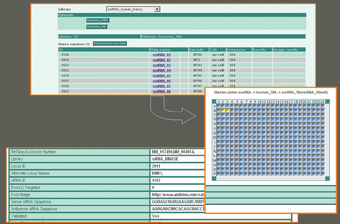

Figure 3: Library browser module – plate viewer
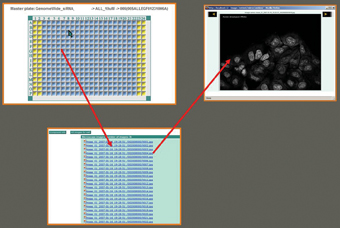

Figure 4: Image viewer
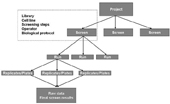

Figure 5: Typical screening hierarchy. Screening parameters are defined on “screen” level and can’t be modified in sublevels
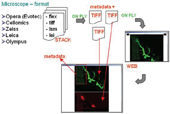

Figure 6: Concept of image converter. By clicking on link (file) or well in plate directly, user can browse in web interface slices and metadata extracted on fly from original microscope file format
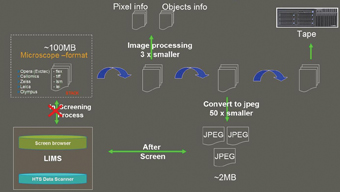

Figure 7: Automated data flow of the HCS facility of the Max Planck Institute in Dresden, Germany
References
- Karol Kozak, Marta Kozak, Jan Wagner, Hannes Grabner, Kerstin Korn, Eugenio Fava, Marit Biesold, Claudia Moebius, Anett Lohman, Ebrhard Krausz: TDS LIMS: a platform for comprehensive management and analysis of screening data. Screening Europe 2006, 20-22 February 2006, http://www.rnai.net/index.aspx?ID=71113
- Karol Kozak, Anne Heninger, Marta Kozak, Ina Poser, Mathias Gierth, Frank Buchholz, Eberhard Krausz: Library Production: a Database for the Enzymatic Production of small interfering RNA (siRNA). Screening Europe 2006, 20-22 February 2006, http://www.rnai.net/ index.aspx?ID=73403
- Karol Kozak, Marta Kozak, Eberhard Krausz: SIB: database and tool for the integration and browsing of large scale image high-throughput screening data. IEEE. Lectures Proceedings. BIDM ’06.
- J. Gallaugher and S. Ramanathan. Choosing a client/server architecture. a comparison of two-tier and three-tier systems. Information Systems Management Magazine, 2(13):7– 13, 1996.
- M. Beveridge, Y. W. Park, J. Hermes, A. Marenghi, G. Brophy, A. Santos, Detection of p56(lck) kinase activity using scintillation proximity assay in 384-well format and imaging proximity assay in 384- and 1536-well format. J. Biomol. Screen. 4, 205-212 (2000).
- Pelkmans, L., Fava, E., Grabner, H., Hannus, M., Habermann, B., Krausz, E., Zerial, M.: Genome-wide analysis of human kinases in clathrin- and caveolae/raft-mediated endocytosis. Nature (2005) 436:78-86.
- Kittler R, Surendranath V, Heninger AK, Slabicki M, Theis M, Putz G, Franke K, Caldarelli A, Grabner H, Kozak K, Wagner J, Rees E, Korn B, Frenzel C, Sachse C, Sonnichsen B, Guo J, Schelter J, Burchard J, Linsley PS, Jackson AL, Habermann B, Buchholz F.: Genome-wide resources of endoribonuclease-prepared short interfering RNAs for specific loss-of-function studies. Nat Methods (2007) 4:337-344.
- High Throughput Technology Development Studio (TDS) at the Max Planck Institute of Molecular Cell Biology and Genetics. http://tds.mpi-cbg.de
- The Center for Information Services and High Performance Computing. http://tu-dresden.de/die_tu_dresden/zentrale_einrichtungen/zih/
- Tomasz J. Proszynski, Robin Klemm, Maike Gravert, Peggy Hsu, Jan Wagner, Karol Kozak, Hannes Grabner, Bianca Habermann, Michel Bagnat, Kai Simons and Christiane Walch-Solimena: A visual screen for sorting mutants in yeast biosynthetic pathways using the systematic deletion mutant array. PNAS (2005) 102:17981-17986.
- W. Zheng, S. S. Carroll, J. Inglese, R. Graves, L. Howells, B. Strulovici, Miniaturization of a hepatitis C virus RNA polymerase assay using a ?102 degrees C cooled CCD camera-based imaging system. Anal. Biochem. 290, 214-220 (2001).
- Krausz, E.: Challenges in High-Content siRNA Screening. European Pharmaceutical Review (2006) (6)13-20.
Karol Kozak
Karol Kozak has been influential in the development of data handling and data mining tools for High Thoroughput/High Content Screening (HCS) at Max Planck institute of Molecular Cell Biology and Gentic in Dresden, (Germany) for a number of years. In the past year he has produced two publications and given five presentations within the HCS sector; as service leader of the data handling facility, he currently plays a leading managerial role in defining the strategy for the organisation of large scale data produced by biologists.




