Is Pharma getting what it needs from RNAi?
Posted: 24 March 2006 | | No comments yet
Large and small drug development companies have used RNAi intensively for several years now1-3. The adoption of RNAi technologies by drug companies followed fairly closely with their adoption by academic research labs, and as such many of the challenges and problems that were a natural consequence of the rapid expansion of RNAi needed to be […]
Large and small drug development companies have used RNAi intensively for several years now1-3. The adoption of RNAi technologies by drug companies followed fairly closely with their adoption by academic research labs, and as such many of the challenges and problems that were a natural consequence of the rapid expansion of RNAi needed to be worked out by the industrial sector along with academia.
This is somewhat unusual for a technological platform. Traditionally, drug development companies have tended to wait until a technology has matured in academia before adopting it, or has looked to small biotechnology companies to establish that a particular technology is robust. With regards to RNAi, biotechnology companies that produced large catalogues of siRNAs have certainly been instrumental in assisting drug companies in establishing RNAi programs, but the implementation of these programs have largely been shouldered by the drug companies themselves. For target validation using siRNAs, these efforts have included developing screening groups and labs, data analysis and a pipeline for validating hits from RNAi screens. For programs that use adenoviral or lentiviral vectors to deliver expressed shRNAs, research labs in industry have established viral containment laboratories and had to implement procedures for working with large numbers of viral constructs. These capital and resource commitments have been significant, but have also facilitated the use of powerful new genetic methods in drug development. However, this process has not been easy.
A changing landscape
Concomitant with the resource and logistical issues of developing in-house RNAi programs, pharmaceutical and biotechnology companies have had to wrestle with complex and vexing complications that arose within RNAi technologies as their use became more widespread. These issues largely revolved around the problem of ‘off-target effects’, or a lack of specificity for an RNAi reagent to hit its intended target without also hitting a significant number of additional genes4. These problems had the potential to severely limit the reliability of RNAi as a useful set of tools for drug discovery. The extent of off-target effects resulting from RNAi reagents became apparent when researchers began interrogating cells treated with RNAi by transcriptional profiling5. These experiments consistently revealed that a large number of genes were affected by any siRNA, and that the set of genes affected changed substantially with each additional siRNA, particularly for siRNAs that target different regions of the same gene. Drug discovery companies, RNAi reagent providers and academic laboratories discussed these effects at length. During this period, roughly from 2002 through 2004, potential causes of off-target effects were examined and rules for minimising them were tested. These efforts were successful in reducing and managing the complications of off-target effects caused by RNAi reagents6 and the urgency surrounding these discussions have largely subsided in the last year.
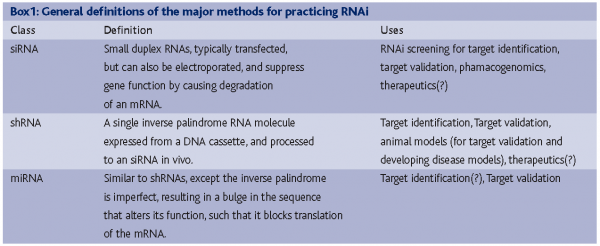
If the technical problems of RNAi have been largely reigned in, where does RNAi stand overall on meeting the high expectations that accompanied the genesis of these research programs? How have the expectations changed for RNAi over the last few years? Questions regarding the success of RNAi will receive different answers from different respondents, but certainly some general trends exist and contrasting viewpoints can be traced to how well specific RNAi technologies fit with specific drug discovery problems. RNAi technologies widely accepted by the industrial sector include siRNAs and shRNAs in the early phases of the drug discovery process, with additional technologies, such as the use of miRNAs for target validation, RNAi screening in model organisms and therapeutic RNAi receiving, are receiving limited, yet still serious, attention. The distinct types and applications of RNAi are described in Box 1. Through 2005, RNAi as an experimental tool consisted of siRNAs7-9 and shRNAs10-14, both of which leverage an endogenous mRNA surveillance pathway that degrades mRNAs selectively. In the last year, an additional endogenous pathway, which uses15,16 to block translation of mRNAs has been better characterised. This pathway uses a set of enzymes that partially overlap with those used by siRNAs to degrade mRNAs, and has been developed as a tool for gene function studies13,17. miRNAs can be expressed exogenously either singly or in clusters to target several genes at once. Several points can be made regarding how these technologies address needs within the drug discovery sector. These needs, and a discussion about how well RNAi is addressing them, can be grouped as follows:
Is RNAi showing success in identifying new drug targets?
One of the immediate promises of RNAi was the ability to screen many genes, and ultimately the entire human genome, on a functional level3,9,18-20. This was regarded as an advance over previous efforts to correlate genes with disease on a large scale, particularly through transcriptional profiling, because screening genes by RNAi could define them as having at least some potential to resolve disease. The identification of genes that induce apoptosis in cancer cell lines, or reduce cytokine expression in epithelial or immune cell lines, using large scale RNAi screens presents a powerful method for identifying new approaches to anticancer and anti-inflammatory therapies8,21-25. Screens for targets within these therapeutic areas have been most frequently performed, owing to the fact that cell lines and assays within oncology and inflammation have been the easiest to format. Additionally, many cell lines within these areas are readily transfectable, and libraries of siRNAs have been faster to develop and test, and this format requires a transfection or electroporation step. In contrast, the most important cell models used in metabolic diseases and neuroscience are difficult to use with siRNAs in a large scale. Additionally, many of the endpoints for these screens require cytological readouts, which have been difficult to scale previously, but can now be addressed by High Content Screening26-28. To expand RNAi screens into underserved therapeutic areas, several companies (Novartis, Bristol Meyers Squibb, Eli Lilly and Merck) have joined a consortium managed by the Broad Institute, the Massachusetts Institute of Technology and Harvard Medical School to develop a library of viral shRNA reagents. Additional viral libraries have been developed, notably from Cold Spring Harbor Laboratories29, and a European project between the Cancer Research unit of the UK Research Institute and the Netherlands Cancer Institute30.
RNAi screens were initially performed in model systems, such as Drosophila31-35 and C. elegans36-39, as these systems were instrumental in characterising RNAi, and the use of RNAi in mammalian cells was only possible following the observations of Elbashir that short duplexes could be transfected into mammalian cells without triggering an endogenous anti-viral response7. This paper triggered the development of large-scale screens from research groups within academia and at many of the major pharmaceutical companies. The first major RNAi screen in mammalian cells was one reported in 2003, which identified genes that suppressed Trail-mediated apoptosis in HeLa cells, by a group at the Genomics Research Institute of the Novartis Foundation25. Following this screen, many additional screens were performed and reported that looked at a wide variety of disease models using both siRNAs40,41 and shRNAs21,22,24,29. These screens have proved to be robust platforms for describing large numbers of genes that can be considered novel therapeutic opportunities, and these screens have opened these new avenues in a very short period of time.
Is RNAi showing success in validating new targets?
Many target validation experiments are technically identical to target identification experiments, in that both explicitly use RNAi to suppress gene function, but they differ in scale. Target validation can occur for several genes simultaneously, but is really a process of carefully evaluating whether a gene identified in a screen truly represents a therapeutic opportunity. These experiments go beyond repeating an experiment, and are intended to expand an initial observation into a package of data to support the next stage of drug development, such as establishing a compound screening campaign. Validation studies have been described for targets in several areas, and while screening has focused largely on oncology and inflammation, target validation studies have been better distributed throughout the major disease areas, including cardiovascular diseases42, diabetes and related disorders43-45, and neurodegeneration46. This is because validation studies are better able to utilise viral methods for introducing RNAi, and therefore complex biological models can be interrogated by RNAi.
There are many caveats to using a phenotype observed in an RNAi screen as an indication that an inhibitor to a catalytic activity of the gene product will achieve a therapeutic effect. An important part of target validation is to address these caveats. Some caveats are conceptual, including the distinctions between how the absence of a protein is a fundamentally different mechanism of inhibiting a process than blocking an activity. Many proteins have several functions and an inhibitor may block only one of them. Other caveats are technical. One of the most important technical concerns is the occurrence of off-target effects, discussed above. However, these effects are becoming less of a concern because target validation groups expect these complications to happen with some regularity, and have developed robust schema to study a candidate target thoroughly before considering a target to be validated. Validation experiments include testing the gene function with several distinct RNAi reagents, careful correlation of a phenotype with the degree of knockdown, and integrating these data with additional approaches, such as expressing dominant negative mutations of the gene in question. Other technical caveats are subtler, such as the significant temporal differences between small molecule inhibitors and RNAi, and even among RNAi treatments of different genes. Again, target validation groups have been able to control for these differences experimentally and analytically.
Is RNAi showing a role in advancing drug candidates?
RNAi is of interest to drug development for reasons that extend beyond the identification and validation of new targets. Making a decision about which compounds will advance into preclinical studies, and the characterisation of a candidate within preclinical studies, is challenging and there are few widely applicable and robust methods available for making these decisions. Again, looking at the extension of transcriptional profiling as a method of identifying genes that correlate with compound effects, towards getting a functional association of genes and compounds using RNAi, it is possible to develop new methods to assist in several key steps of the drug development process. Transcriptional profiling has been used to characterise compound relationships biologically47-49 and these data have been used in conjunction with chemical and pharmacokinetic data to make decisions on which compounds to advance. RNAi has been used in studies that can identify interactions between specific genes and compound sensitivity or toxicity41,50. Examples include screens to identify genes that exacerbate the effects of a drug applied at concentrations below its normal effective concentration. These ‘chemosensitisation’ screens can identify pathways that impact on the effect of a therapy, which may serve as an indication of genetic differences that can increase the efficacy, the toxicity or the resistance to a potential therapeutic (depending on how the screen is configured). These data can be highly informative about how the potential therapeutic functions in a wide set of conditions, potentially alerting a drug development team to issues of resistance and toxicity, as well as help identify compounds that may work in combinations with other drugs.
Is RNAi showing success in becoming useful as a therapeutic?
Interest in developing sequence-specific therapeutics has been an ideal therapeutic strategy for decades. The value of having such a genetic therapeutic succeed is tremendous, as it would expand the definition of a druggable target to include most or all of the human genome. Technologies such as DNA antisense and therapeutic ribozymes have been experimented with, and have even advanced into clinical trials. These approaches have had limited success at best, in partly due to difficulties with pharmacokinetics and efficacy at the preclinical and clinical levels. As such, these types of therapeutics have been developed for a few indications only.
Recently, the success of RNAi has peaked in interest as a therapeutic with the potential to perform more reliably and without the toxicities observed with previous approaches51,52. Interest exists for both siRNAs53, and shRNAs expressed from viral vectors54,55, as potential therapeutics. However, the latter is operationally identical to gene therapy approaches that seek to replace mutationally inactivated genes for inherited disorders such as adenosine deaminase deficiency and p53 for some cancers. These approaches are limited by significant technical challenges involving the ability to introduce genetic information into patients safely. In contrast, the use of siRNAs for therapeutic benefit is being actively pursued by several significant collaborations between biotechnology companies and large pharmaceutical companies, including a collaboration between Sirna and Eli Lilly, and collaborations between Alnylam and Merck and Novartis.
Making the most of RNAi
In conclusion, it is fair to say that pharma is getting many of the things that it needs from RNAi. Those things for which RNAi has not yet delivered are commanding considerable attention from both pharmaceutical and biotechnology companies. Specifically, the early phases of the drug discovery pipeline have undergone important changes in they way they are practiced, with more information on a larger set of potential targets readily available. The utility of RNAi in the later phases of preclinical research is still under development, but may be important additions to selecting clinical candidates and using them effectively in clinical settings. Therapeutic applications of RNAi represent the most dramatic benefit to the drug development and medicine, but it will be several years before any real clarity will emerge on what its real contributions can be.

References
- RNA Interference Technology: From Basic Science to Drug Development. 1st ed, ed. K. Appasani. 2005, Cambridge, UK: Cambridge University Press. 510.
- Voorhoeve, P.M. and R. Agami, Knockdown stands up. Trends Biotechnol 2003; 21(1); 2-4.
- Willingham, A.T., Q.L. Deveraux, G.M. Hampton, and P. Aza-Blanc, RNAi and HTS: exploring cancer by systematic loss-of-function. Oncogene 2004; 23; 8392-8400.
- Jackson, A.L. and P.S. Linsley, Noise amidst the silence: off-target effects of siRNAs? Trends in Genetics 2004; 20; 521-524.
- Jackson, A.L., S.R. Bartz, J. Schelter, S.V. Kobayashi, J. Burchard, M. Mao, B. Li, G. Cavet, and P.S. Linsley, Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol 2003; 21; 635-637.
- Huppi, K., S.E. Martin, and N.J. Caplen, Defining and Assaying RNAi in Mammalian Cells. Molecular Cell 2005; 17; 1-10.
- Elbashir, S.M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl, Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001; 411(6836); 494-8.
- Erfle, H., J.C. Simpson, P.I. Bastiaens, and R. Pepperkok, siRNA cell arrays for high-content screening microscopy. Biotechniques 2004; 37; 454-458.
- Sachse, C. and C.J. Echeverri, Oncology studies using siRNA libraries: the dawn of RNAi-based genomics. Oncogene 2004; 23; 8384-8391.
- Brummelkamp, T.R., R. Bernards, and R. Agami, A system for stable expression of short interfering RNAs in mammalian cells. Science 2002; 296(5567); 550-3.
- Paul, C.P., P.D. Good, I. Winer, and D.R. Engelke, Effective expression of small interfering RNA in human cells. Nat Biotechnol 2002; 20(5); 505-8.
- Yu, J.Y., S.L. DeRuiter, and D.L. Turner, RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc Natl Acad Sci U S A 2002; 99(9); 6047-52.
- McManus, M.T., C.P. Petersen, B.B. Haines, J. Chen, and P.A. Sharp, Gene silencing using micro-RNA designed hairpins. Rna 2002; 8(6); 842-50.
- Miyagishi, M. and K. Taira, U6 promoter-driven siRNAs with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat Biotechnol 2002; 20(5); 497-500.
- Carrington, J.C. and V. Ambros, Role of microRNAs in plant and animal development. Science 2003; 301; 336-338.
- Du, T. and P.D. Zamore, microPrimer: the biogenesis and function of microRNA. Development 2005; 132; 4645-4652.
- Zeng, Y., X. Cai, and B.R. Cullen, Use of RNA Polymerase II to Transcribe Artificial MicroRNAs, in Methods of Enzymology, D.R. Engelke and J.J. Rossi, Editors. 2005, Academic Press: New York, NY. p. 371-380.
- Downward, J., Use of RNA interference libraries to investigate oncogenic signalling in mammalian cells. Oncogene 2004; 23; 8376-8383.
- O’Neil, N.J., R.L. Martin, M.L. Tomlinson, M.R. Jones, A. Coulson, and P.E. Kuwabara, RNA-mediated interference as a tool for identifying drug targets. Am J Pharmacogenomics 2001; 1(1); 45-53.
- Wheeler, D.B., S.N. Bailey, D.A. Guertin, A.E. Carpenter, C.O. Higgins, and D.M. Sabatini, RNAi living-cell microarrays for loss-of-function screens in Drosophila melanogaster cells. Nat Methods 2004; 1; 127-32.
- Berns, K., E.M. Hijmans, J. Mullenders, T.R. Brummelkamp, A. Velds, M. Heimerikx, R.M. Kerkhoven, M. Madiredjo, W. Nijkamp, B. Weigelt, R. Agami, W. Ge, G. Cavet, P.S. Linsley, R.L. Beijersbergen, and R. Bernards, A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature 2004; 428; 431-437.
- Kolfschoten, I.G., B. van Leeuwen, K. Berns, J. Mullenders, R.L. Beijersbergen, R. Bernards, P.M. Voorhoeve, and R. Agami, A genetic screen identifies PITX1 as a suppressor of RAS activity and tumorigenicity. Cell 2005; 121; 849-858.
- MacKeigan, J.P., L.O. Murphy, and J. Blenis, Sensitized RNAi screen of human kinases and phosphatases identifies new regulators of apoptosis and chemoresistance. Nature Cell Biology 2005; 7; 591-600.
- Westbrook, T.F., E.S. Martin, M.R. Schlabach, Y. Leng, A.C. Liang, B. Feng, J.J. Zhao, T.M. Roberts, G. Mandel, G.J. Hannon, R.A. Depinho, L. Chin, and S.J. Elledge, A genetic screen for candidate tumor suppressors identifies REST. Cell 2005; 121; 837-848.
- Aza-Blanc, P., C.L. Cooper, K. Wagner, S. Batalov, Q.L. Deveraux, and M.P. Cooke, Identification of modulators of TRAIL-induced apoptosis via RNAi- based phenotypic screening. Molecular Cell 2003; 12; 627-637.
- Haney, S.A., RNAi and high content screening in target identification and validation. IDrugs 2005; 8; 997-1001.
- Comley, J., High content screening-Emerging importance of novel reagents/probes and pathway analysis. Drug Discovery World 2005; 6(31-54).
- Rausch, O., Use of high-content analysis for compound screening and target selection. IDrugs 2005; 8; 573-577.
- Paddison, P.J., J.M. Silva, D.S. Conklin, M. Schlabach, M. Li, S. Aruleba, V. Balija, A. O’Shaughnessy, L. Gnoj, K. Scobie, K. Chang, T. Westbrook, M. Cleary, R. Sachidanandam, W.R. McCombie, S.J. Elledge, and G.J. Hannon, A resource for large-scale RNA-interference-based screens in mammals. Nature 2004; 428; 427-431.
- Frankish, H., Consortium uses RNAi to uncover genes’ function. Lancet 2003; 361; 584.
- Kiger, A.A., B. Baum, S. Jones, M.R. Jones, A. Coulson, C. Echeverri, and N. Perrimon, A functional genomic analysis of cell morphology using RNA interference. Journal of Biology 2003; 2; 27.
- Caplen, N.J., J. Fleenor, A. Fire, and R.A. Morgan, dsRNA-mediated gene silencing in cultured Drosophila cells: a tissue culture model for the analysis of RNA interference. Gene 2000; 252; 95-105.
- Lum, L., S. Yao, B. Mozer, A. Rovescalli, D. Von Kessler, M. Nirenberg, and P.A. Beachy, Identification of Hedgehog pathway components by RNAi in Drosophila cultured cells. Science 2003; 299; 2039-2045.
- Boutros, M., A.A. Kiger, S. Armknecht, K. Kerr, M. Hild, B. Koch, S.A. Haas, H.F. Consortium, R. Paro, and N. Perrimon, Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science 2004; 303; 832-835.
- Kim, Y.O., S.J. Park, R.S. Balaban, M. Nirenberg, and Y. Kim, A functional genomic screen for cardiogenic genes using RNA interference in developing Drosophila embryos. Proc Natl Acad Sci U S A 2004; 101; 159-164.
- Colaiacovo, M.P., G.M. Stanfield, K.C. Reddy, C.M. Reinke, Stanfield GM, Reddy KC, Reinke V., S.K. Kim, and A.M. Villeneuve, A targeted RNAi screen for genes involved in chromosome morphogenesis and nuclear organization in the Caenorhabditis elegans germline. Genetics 2002; 162; 113-128.
- Lee, S.S., R.Y. Lee, A.G. Fraser, R.S. Kamath, J. Ahringer, and G. Ruvkun, A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nature Genetics 2003; 33; 40-48.
- Pothof, J., G. van Haaften, K. Thijssen, R.S. Kamath, A.G. Fraser, J. Ahringer, R.H. Plasterk, and M. Tijsterman, Identification of genes that protect the C. elegans genome against mutations by genome-wide RNAi. Genes and Development 2003; 17; 443-448.
- Sonnichsen, B., L.B. Koski, A. Walsh, P. Marschall, B. Neumann, M. Brehm, A.M. Alleaume, J. Artelt, P. Bettencourt, E. Cassin, M. Hewitson, C. Holz, M. Khan, S. Lazik, C. Martin, B. Nitzsche, M. Ruer, J. Stamford, M. Winzi, R. Heinkel, M. Roder, J. Finell, H. Hantsch, S.J. Jones, M. Jones, F. Piano, K.C. Gunsalus, K. Oegema, P. Gonczy, A. Coulson, A.A. Hyman, and C.J. Echeverri, Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 2005; 434; 462-469.
- Kittler, R., G. Putz, L. Pelletier, I. Poser, A.K. Heninger, D. Drechsel, S. Fischer, I. Konstantinova, B. Habermann, H. Grabner, M.L. Yaspo, H. Himmelbauer, B. Korn, K. Neugebauer, M.T. Pisabarro, and F. Buchholz, An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature 2004; 432; 1036-1040.
- MacKeigan, J.P., L.O. Murphy, and B. J., Sensitized RNAi screen of human kinases and phosphatases identifies new regulators of apoptosis and chemoresistance. Nature Cell Biology 2005; 7; 591-600.
- Kasahara, H. and H. Aoki, Gene silencing using adenoviral RNAi vector in vascular smooth muscle cells and cardiomyocytes. Methods in Molecular Medicine 2005; 112; 155-172.
- Pirola, L., S. Bonnafous, A.M. Johnston, C. Chaussade, F. Portis, and E. Van Obberghen, Phosphoinositide 3-kinase-mediated reduction of insulin receptor substrate-1/2 protein expression via different mechanisms contributes to the insulin-induced desensitization of its signaling pathways in L6 muscle cells. Journal of Biological Chemistry 2003; 278; 15641-15651.
- Taniguchi, C.M., i.K. Uek, and R. Kahn, Complementary roles of IRS-1 and IRS-2 in the hepatic regulation of metabolism. Journal of Clinical Investigation 2005; 115; 718-727.
- Oshitari, T., D. Brown, and S. Roy, SiRNA strategy against overexpression of extracellular matrix in diabetic retinopathy. Experimental Eye Research 2005; 81; 32-37.
- Thakker, D.R., D. Hoyer, and J.F. Cryan, Interfering with the brain: Use of RNA interference for understanding the pathophysiology of psychiatric and neurological disorders. Pharmacology and Therapeutics 2006; 109; 413-438.
- Clarke, P.A., R. te Poele, R. Wooster, and P. Workman, Gene expression microarray analysis in cancer biology, pharmacology, and drug development: progress and potential. Biochemical Pharmacology 2001; 62; 1311-1336.
- Butcher, R.A. and S.L. Schreiber, Using genome-wide transcriptional profiling to elucidate small-molecule mechanism. Current Opinion in Chemical Biology 2005; 9; 25-30.
- Heuser, M., L.U. Wingen, D. Steinemann, G. Cario, N. von Neuhoff, M. Tauscher, L. Bullinger, J. Krauter, G. Heil, H. Dohner, B. Schlegelberger, and A. Ganser, Gene-expression profiles and their association with drug resistance in adult acute myeloid leukemia. Haematologica 2005; 90; 1484-1492.
- Mousses, S., A. Kallioniemi, P. Kauraniemi, A. Elkahloun, and O.-P. Kallioniemi, Clinical and functional target validation using tissue and cell microarrays. Current Opinion in Chemical Biology 2001; 6; 97-101.
- Shankar, P., N. Manjunath, and J. Lieberman, The Prospect of Silencing Disease Using RNA Interference. JAMA 2005; 293; 1367-1373.
- Stevenson, M., Therapeutic potential of RNA interference. New England Journal of Medicine. 2004; 351; 1772-1777.
- Leung, R.K. and P.A. Whittaker, RNA interference: from gene silencing to gene-specific therapeutics. Pharmacology and Therapeutics 2005; 107; 222-239.
- Xuan, B., Z. Qian, J. Hong, and W. Huang, EsiRNAs inhibit Hepatitis B virus replication in mice model more efficiently than synthesized siRNAs. Virus Research 2006; in press.
- Grimm, D. and M.A. Kay, Therapeutic short hairpin RNA expression in the liver: viral targets and vectors. Gene Therapy 2006; in press.
About the author – Steven A. Haney
Dr Haney received his Ph.D. from the Department of Biological Chemistry at the University of Michigan. Following his graduate studies, he was a Research Fellow in the Department of Molecular Biology at Princeton University. His post-doctoral work focused on the genetic analysis of Ras signaling in yeast. In 1995, Dr Haney joined Cadus pharmaceuticals developing systems for the study of human GPCRs through genetic systems developed in yeast and human cell lines. From there he went to the Department of Infectious Diseases in 1998, where he adapted yeast genetic systems for the identification and characterisation of protein-protein interaction based drug targets in bacterial and viral pathogens. These studies led to the development of drug discovery programs that successfully identified inhibitors of a protein-protein interaction. In 2001 Dr Haney joined the Genomics Department at Wyeth Research, developing platform-based methods for target identification and validation to treat diseases in cancer and inflammation. Recently, his group has focused cancers of several tissues and has employed cell biological methods, especially High Content Screening, for the study of genes and compounds affecting cancer cell growth.




