Drug screens on human stem cells: From understanding cell biology to predicting drug toxicity
Posted: 19 October 2011 |
The Canadian physician William Osler said: “The person who takes medicine must recover twice, once from the disease and once from the medicine.” Indeed, all medicines have side effects – some of which may complicate a patient’s treatment, or in extreme cases may even be fatal. Of concern is the fact that side effects of drugs are often difficult or impossible to predict from preclinical studies, and infamous cases of drugs causing permanent injury or even death of patients in clinical trials illustrate the severity of this problem.
Late stage attrition of drugs is costly for pharmaceutical companies, contributing to the rise of drug prices and delays of drug delivery to market. Better prediction of compound efficacy and safety at early stages of drug development relies on improvement of the models used for pre-clinical testing. The availability of human embryonic stem (hES) cells and, more recently, induced pluripotent stem (iPS) cells may transform the landscape of drug discovery. Here we provide an overview of human pluripotent stem cell features that make them amenable to predictive toxicology and discuss how chemical screens that aim to find drugs that modulate stem cell fates may provide a paradigm for using stem cells in drug discovery.
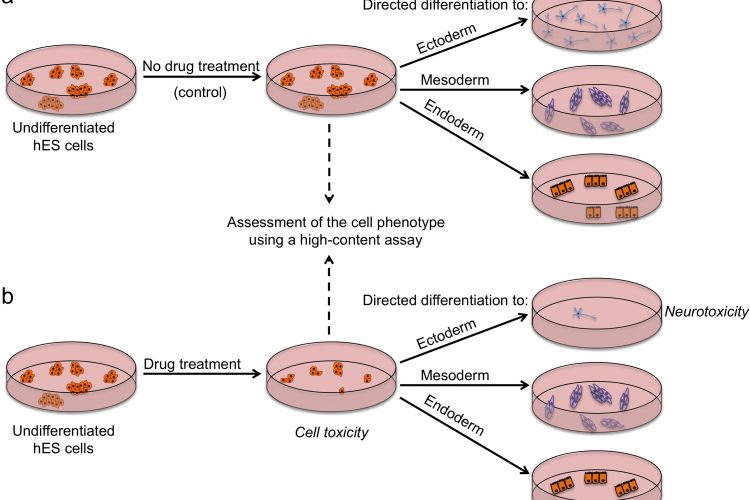

FIGURE 2 Outline of a strategy for testing drugs that perturb embryonic development. Undifferentiated hES cells are grown in (a) control conditions and (b) in the presence of a drug, and then assessed by a high content assay to determine the effect of a drug on cells (by examining the number of cells, expression of markers associated with the differentiated and undifferentiated state, colony number and size). Cells from both conditions are then induced to differentiate to specific cell types to assess the effect of drug treatment on the differentiation ability of hES cells. For example, low number of neurons upon drug treatment may indicate neurotoxic effects of the drug
The Canadian physician William Osler said: “The person who takes medicine must recover twice, once from the disease and once from the medicine.” Indeed, all medicines have side effects – some of which may complicate a patient’s treatment, or in extreme cases may even be fatal. Of concern is the fact that side effects of drugs are often difficult or impossible to predict from preclinical studies, and infamous cases of drugs causing permanent injury or even death of patients in clinical trials illustrate the severity of this problem.
Late stage attrition of drugs is costly for pharmaceutical companies, contributing to the rise of drug prices and delays of drug delivery to market. Better prediction of compound efficacy and safety at early stages of drug development relies on improvement of the models used for pre-clinical testing. The availability of human embryonic stem (hES) cells and, more recently, induced pluripotent stem (iPS) cells may transform the landscape of drug discovery. Here we provide an overview of human pluripotent stem cell features that make them amenable to predictive toxicology and discuss how chemical screens that aim to find drugs that modulate stem cell fates may provide a paradigm for using stem cells in drug discovery.
Characteristics of human pluripotent stem cell lines
Human ES cells are obtained from the inner cell mass of surplus blastocysts that have been created by in vitro fertilisation for the purpose of assisted reproduction1. When placed under appropriate culture conditions, these karyotypically normal, untransformed cells can be propagated in an undifferentiated state for prolonged periods, seemingly indefinitely, due to their unlimited capacity for self-renewal. Importantly, even after extensive propagation in vitro, they still retain the ability to differentiate into derivatives of all three embryonic germ layers (pluripotency). Human ES cells have a distinct molecular profile defined by expression of a characteristic set of cell-surface and intracellular antigens. Cell surface antigens found in high levels on hES cells include globoseries glycolipid antigens SSEA3 and SSEA4, and the keratan sulfate antigens TRA-1-60 and TRA-1-812. Transcription factors OCT4 (POU5F1) and NANOG are also associated with the undifferentiated state2.
The progress of hES cell research has been controversial and hampered by ethical issues concerning the destruction of human blastocysts during the process of ES cell derivation. Thus, the discovery of a method of generating pluripotent cells very similar to ES cells by reprogramming somatic cells has been met with great enthusiasm. In breakthrough experiments, Yamanaka and colleagues reprogrammed first mouse (2006) then human fibroblast cells (2007) using overexpression of just four transcription factors: Oct-4, Sox2, Klf4 and c-Myc3,4. The simplicity of the method and its great potential benefits resonated with the scientific community, and numerous studies over the last five years have embraced this technology. In addition to providing a scientific tool to study fundamental questions about developmental plasticity and reprogramming, iPS cells offer a means of obtaining patientspecific cells that could be used for disease modelling and drug screens.
Drug screens on stem cells: using drugs to uncover stem cell mechanisms
The alternative potential fates of any pluripotent stem cell in culture are self-renewal, diff – erentiation or death. The main challenge for stem cell scientists is controlling the processes that govern these fate ‘decisions’ in order to: (i) achieve expansion of undifferentiated ES/iPS cells in an effective and robust way, and (ii) direct ES/iPS cells to differentiate into desired functional cell types, such as cardiomyocytes or hepatocytes. Low molecular weight compounds are proving particularly useful in these endeavours. They can be simply applied to media and their effect can be titrated by varying the compound concentration.
Promoting cell survival with small molecules
Current culture conditions for maintenance of undifferentiated hES cells are clearly suboptimal as they result in a high cell death rate. Based on the fact that hES cells have a cell cycle time of less than 24 hours and are routinely split every four or five days at only 1:2 or 1:3 ratio, Olariu et al. estimated that as many as 90 per cent of the potential yield of cells are lost between passages5. This means that not only is it difficult to obtain large numbers of ES cells, but it also creates selection pressure in culture. Under sub-optimal conditions, a spontaneous genetic change or changes that confer a growth advantage on a cell is likely to lead to this cell type expanding and outcompeting its normal counterparts. Indeed, cells with consistent types of karyotypic changes have been observed in hES cell cultures, and such cells are considered culture-adapted and no longer normal6,7. The issue of culture adaptation may have implications for the use of hES cells in therapies as this process may confer malignant properties on the cells8. Thus, approaches to optimise culture conditions in order to promote survival of ES cells come sharply into focus.
Screening of chemical libraries with the aim of identifying compounds that promote hES cell survival have yielded excellent candidates (Table 1), some of which are now included in commercial media for hES/iPS cell maintenance. In a pioneering study, Watanabe et al. reported the striking effect of the Rho-associated coiled coil kinase (ROCK) inhibitor Y-27632 in increasing the cloning efficiency of hES cells9. Interestingly, most of compounds that were subsequently identified as pro-survival compounds in high-throughput screens on hES cells by our group and others also turned out to be inhibitors of ROCK (Table 1). An important lesson about varying drug actions in different types of cells is exemplified by the FDAapproved drug pinacidil, identified in our screen as a pro-survival compound in hES cells. Pinacidil is an opener of KATP-channels in various cell lines and primary human cells10,11. However, it appears to exert its effects on ES cells not through this mechanism, but by inhibition of ROCK12.
Table 1: Examples of pro-survival small compounds identified in chemical screens
| Compound | Target(s) | Ref. |
| Y-27632 | ROCK | 9 |
| Pinacidil | ROCK, PRK2, RSK1, MNK1 | 12 |
| HA1077 | ROCK, PRK2, RSK1, MNK1 | 12,27 |
| H-89 | ROCK, PRK2, RSK1, MNK1 | 12 |
| HA1004 | PKA | 12 |
| Thiazovivin | ROCK | 28 |
| Tyrintegin | ROCK | 28 |
| DDD00033325 | ROCK, PRK2 | 29 |
Differentiating stem cells with small molecules
Conventional protocols for differentiation rely on the spontaneous tendency of ES cells to differentiate upon removal of self-renewal factors from the medium, or by inducing the formation of three-dimensional embryoid bodies. Although various cell types from all three embryonic germ layers can be detected upon spontaneous differentiation of ES cells, this haphazard process is inefficient in creating a particular cell type of choice. Implementation of cell-based screening approaches greatly facilitated discovery of novel protocols for differentiation (Table 2). Cells are treated with collections of hundreds or thousands of compounds followed by analysis to detect compounds that give rise to a phenotype of interest (e.g. a presence of lineage-specific or cell-type-specific marker(s)). Phenotype-based screens do not make a priori assumptions about signalling pathways involved and are thus useful in uncovering novel pathways that contribute to stem cell fate. In a screen of 4,000 compounds on mouse ES cells, Borowiak et al. identified molecules (IDE1 and IDE2) that induced definitive endoderm differentiation in up to 80 per cent of cells, revealing a useful alternative to the expensive recombinant Activin A molecule usually used for promoting endoderm differentiation13. A similar screen by Zhu et al. also looked for inducers of endoderm differentiation but in the presence of a low concentration of Activin A. A compound identified in their screen, stauprimide, could not induce endoderm differentiation in the absence of Activin A, but it was a potent enhancer of directed differentiation, not just to endoderm, but also to mesodermal and neuronal lineages14.
Table 2. Examples of differentiation-inducing/promoting small compounds identified in chemical screens.
| Compound | Screening strategy | Effect | Ref. |
| Ascorbic acid | Microscopic observation of aMHC-eGFP reporter mouse ES cell line | Enhances differentiation of mouse ES cells to cardiac myocytes | 30 |
| Cardiogenol (A-D) | Plate reader-based assay using ANF-Luc reporter mouse embryonal carcinoma cells | Induce cardiomyogenesis in mouse ES cells | 31 |
| IDE1/2 | High-throughput FACS assay using Sox17-dsRed reporter mouse ES cell line | Induce definitive endoderm differentiation in mouse and hES cells | 13 |
| Indolactam V | High-content imaging of PDX1-immunolabelled mouse ES cells | Induces differentiation of hES cell-derived endoderm progenitors to pancreatic lineage | 32 |
| Stauprimide | High-content imaging of Sox17-immunolabelled mouse ES cells | Enhances directed differentiation in mouse and hES cells | 14 |
| TWS119 | Plate reader-based assay using Ta1-tubulin-Luc reporter mouse embryonal carcinoma cells | Induces neurogenesis in mouse ES cells | 33 |
Although high-throughput screens on ES cells demonstrated success in identifying small molecule regulators of differentiation, the hits identified mainly enhanced the proportion of cells committed to differentiation or in the best cases resulted in partially differentiated cells, rather than produced terminally differentiated cells. This is perhaps not surprising, especially in relatively small-scale screens, as it is unlikely for a single molecule to affect all the relevant targets or trigger all the signalling cascades that are required to achieve a terminally differentiated state. During development, cells are exposed to a plethora of carefully balanced signalling molecules that guide them through this process. Novel approaches, such as the Combinatorial Cell Culture approach used by Plasticell (www.plasticell.co.uk) that combine a sequential differentiation approach with screening of random compounds15 are expected to further facilitate discovery of the sequential conditions required for differentiation of stem cells to a variety of cell types.
Regardless of the screening approach used, a robust and reliable assay is the crux of phenotype-based screens. In the past decade, high-content screening platforms have become the method of choice for phenotype-based screens12,16,17. High-content screening is often based on automated microscopy that enables rapid image acquisition of fluorescentlylabelled cells, while associated image analysis software allows concomitant analysis of multiple parameters at the single cell level. Our high-content assay for chemical screening on hES cells assessed the effect of compounds on the number of cells in a colony, colony area and shape, intensity of nuclear staining, the percentage of cells in the colony that expressed a marker of pluripotency (TRA-1-60), as well as the number of colonies per well. Importantly, a change in any of these parameters may reflect a perturbation of the stem cell state by a compound12.
Drug screens on stem cells: using stem cells to uncover drug mechanisms
Chemical screening efforts to uncover key signalling networks in ES cells have demonstrated the suitability of ES cells as a screening platform to identify drugs that perturb the stem cell fates. These studies were mainly performed with the aim of finding drugs for manipulating stem cell fates, but they may pave the way for development of improved preclinical assays for testing drug safety. Current methods for testing toxicity of drugs prior to clinical trials involve animal testing which is expensive and slow. Furthermore, interspecies differences may contribute to failure to detect factors that could affect human health18. Humanised in vitro models offer an attractive possibility to avoid animal use. The unique properties of human pluripotent stem cells offer several advantages over the currently used cellbased models for testing drug safety (Figure 1).
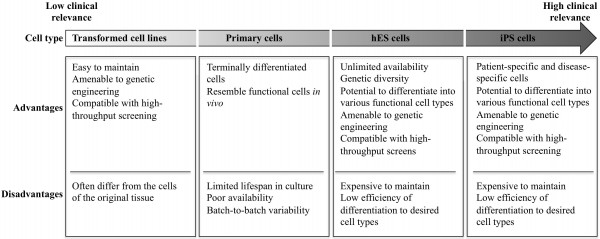

FIGURE 1 Comparison of in vitro models for drug discovery
There are several strategies by which pluripotent stem cells can be used for testing drug safety. First, they could be used in an undifferentiated state to assess the effects of drugs on stem cells and their subsequent ability to differentiate (Figure 2). Tests on undifferentiated cells may be particularly useful for predicting developmental toxicity as developing human embryos are exceptionally vulnerable to drugs and toxic environmental agents. Drugs that are safe to be used by adults may cause defects in prenatal development, and a pregnant woman is most likely to take potentially harmful drugs in the first few weeks post-conception, before she is aware she is pregnant. Of particular concern in testing for effects on development is that animal embryos used for testing may not respond to drugs in the same way as human ones and that it is difficult to gain sufficient data on embryos across a range of concentrations. Thus, access to early embryonic cells of human origin might be particularly helpful in determining drugs affecting developmental pathways. Although no extensive validation studies have been done so far, Adler et al. provided a proof-of-concept study which involved testing a set of known embryotoxicants on hES cells, hES cell-derived progenitors and human foreskin fibroblasts and found that all-trans retinoic acid and 13-cis retinoic acid were more cytotoxic to hES and hES-cell derived cells than fibroblasts19. It is important to note that 13-cis retinoic acid is highly teratogenic in human embryos compared to mouse or rat, highlighting again the issue of interspecies differences and the need for human models20. Despite showing encouraging results, it is clear that the assays used have to be more extensive if they are to address the issue of drugs perturbing developmental processes. Undoubtedly, the more encompassing the screen, the more likely it is to detect potentially unanticipated effects of drugs. High-content assays that we and others used in screens to identify chemicals that affect stem cell fates could be modified and applied here with the purpose of extensive characterisation of stem cells after drug treatment12,16 17. Multiple readouts including cell numbers, cell morphology, presence / absence of lineage-specific markers and the level of their expression would enable quantitative assessment of stem cell fate perturbations by a drug. Further development of this assay could include making pathway-specific or lineagespecific reporter cell lines that could be monitored in real-time as cells are guided through specific differentiation processes, using established protocols for directed diff – erentiation21. A particular advantage of this model in comparison to whole-embryo animal assays is that it allows mechanistic insights into the developmental pathways altered and should help define the timing, length and dose of drug exposure harmful for development.
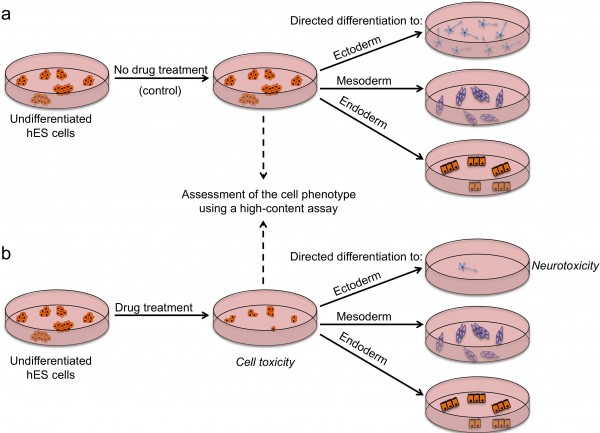

FIGURE 2 Outline of a strategy for testing drugs that perturb embryonic development. Undifferentiated hES cells are grown in (a) control conditions and (b) in the presence of a drug, and then assessed by a high content assay to determine the effect of a drug on cells (by examining the number of cells, expression of markers associated with the differentiated and undifferentiated state, colony number and size). Cells from both conditions are then induced to differentiate to specific cell types to assess the effect of drug treatment on the differentiation ability of hES cells. For example, low number of neurons upon drug treatment may indicate neurotoxic effects of the drug
A second potential strategy for using pluripotent stem cells in drug safety testing entails producing hES/iPS cell-derived differentiated cells, and testing the effect of drugs on particular functional cell types. Given that cardiotoxicity and hepatotoxicity are the main reasons for withdrawal of drugs from late stages of development or after market release, significant efforts have been directed towards developing reliable in vitro assays that could predict adverse effects of drugs on cardiac and liver tissues. The major obstacle in these pursuits is the availability of relevant primary human cells. Ability to derive cardiomyocytes or hepatocytes in vitro from an unlimited supply of undifferentiated pluripotent stem cells will facilitate preclinical testing. Sartipy and Bjorquist have recently reviewed encouraging reports of the application of hES/iPS cell-derived cardiomyocytes and hepatocytes in drug discovery22. Despite the progress in this field, several key challenges remain to be addressed: (i) further improvement of differentiation protocols is pivotal for achieving efficient and robust differentiation that could be scaled up to the high-throughput requirements of the pharmaceutical industry; (ii) differentiated cells derived from hES/iPS cells must be extensively characterised to demonstrate their functional attributes and similarities to their in vivo counterparts; and (iii) assays used for predictive toxicology must also be validated (e.g. using known drugs) to confirm that in vitro effects do indeed correlate with the in vivo effects of drugs.
Finally, the advent of iPS cell technology offers the potential to test drugs on disease–relevant cells. An instructive example is that by Ebert et al. who showed that iPS cells can mimic some aspects of a disease phenotype in vitro23. Fibroblasts of a patient with spinal muscular atrophy were reprogrammed to iPS cells and then induced to differentiate to motor neurons. The motor neuron production was selectively reduced when the patient’s iPS cells were used compared to the wild type iPS cells obtained from his healthy mother, and the diseased motor neurons showed changes in size and expression of synapsin. Interestingly, the authors then used iPS cell-derived motor neurons to test known drugs that ameliorate the disease phenotype in other culture systems. A similar approach for validating candidate drugs was also used for iPS cell-derived cardiomyocytes from patients with Long QT syndrome24,25 and iPS cell-derived neurons from patients with familial dysautonomia26, illustrating the suitability of diseased iPS cells to drug discovery and testing.
Conclusion and future prospects
The growing interest of the pharmaceutical industry in applying hES/iPS cells and their differentiated derivatives in preclinical testing stems from the fact that currently used models are plagued with pitfalls that result in poor prediction of drug safety and efficacy. Although hES/iPS cells hold great promise for improving these tests, significant challenges still have to be met, particularly in optimising protocols for cell maintenance and differentiation. Furthermore, the ultimate goal of designing robust stem cell-based in vitro assays that can accurately predict drug safety and efficacy will require a multidisciplinary approach and good collaboration between stem cell biologists and pharmacologists. Collaborative initiatives between academia and industry, such as the Stem Cells 4 Safer Medicine programme in the UK (www.sc4sm.org), aim to provide a platform for such collaborations. Given the tremendous progress in the field of human pluripotent stem cells achieved in the last decade, there is every reason for optimism that the approaches discussed in this review will result in improved assays for drug discovery.
References
1. Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, et al.: Embryonic stem cell lines derived from human blastocysts. Science 1998; 282:1145-1147
2. Adewumi O, Aflatoonian B, Ahrlund-Richter L, Amit M, Andrews PW, Beighton G, et al.: Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol 2007; 25:803-816
3. Takahashi K, Yamanaka S: Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006; 126:663-676.
4. Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al.: Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007; 131:861-872
5. Olariu V, Coca D, Billings SA, Tonge P, Gokhale P, Andrews PW, et al.: Modified variational Bayes EM estimation of hidden Markov tree model of cell lineages. Bioinformatics 2009; 25:2824-2830
6. Draper JS, Smith K, Gokhale P, Moore HD, Maltby E, Johnson J, et al.: Recurrent gain of chromosomes 17q and 12 in cultured human embryonic stem cells. Nat Biotechnol 2004; 22:53-54
7. Baker DE, Harrison NJ, Maltby E, Smith K, Moore HD, Shaw PJ, et al.: Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat Biotechnol 2007; 25:207-215
8. Ben-David U, Benvenisty N: The tumorigenicity of human embryonic and induced pluripotent stem cells. Nat Rev Cancer 2011; 11:268-277
9. Watanabe K, Ueno M, Kamiya D, Nishiyama A, Matsumura M, Wataya T, et al.: A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat Biotechnol 2007; 25:681-686
10. Arrigoni-Martelli E, Nielsen CK, Olsen UB, Petersen HJ: N”-cyano-N-4-pyridyl-N’-1,2,2- trimethylpropylguanidine, monohydrate (P 1134): a new, potent vasodilator. Experientia 1980; 36:445-447
11. Arena JP, Kass RS: Enhancement of potassiumsensitive current in heart cells by pinacidil. Evidence for modulation of the ATP-sensitive potassium channel. Circ Res 1989; 65:436-445
12. Barbaric I, Gokhale PJ, Jones M, Glen A, Baker D, Andrews PW: Novel regulators of stem cell fates identified by a multivariate phenotype screen of small compounds on human embryonic stem cell colonies. Stem Cell Res 2010; 5:104-119
13. Borowiak M, Maehr R, Chen S, Chen AE, Tang W, Fox JL, et al.: Small molecules efficiently direct endodermal differentiation of mouse and human embryonic stem cells. Cell Stem Cell 2009; 4:348-358
14. Zhu S, Wurdak H, Wang J, Lyssiotis CA, Peters EC, Cho CY, et al.: A small molecule primes embryonic stem cells for differentiation. Cell Stem Cell 2009; 4:416-426
15. Choo Y: Use of Combinatorial Screening to Discover Protocols that Effectively Direct the Differentiation of Stem Cells. In Shi Y CD (ed): Stem Cell Research and Therapeutics: Springer, 2008:227-250
16. Barbaric I, Gokhale PJ, Andrews PW: High-content screening of small compounds on human embryonic stem cells. Biochem Soc Trans 2010; 38:1046-1050.
17. Barbaric I, Jones M, Harley DJ, Gokhale PJ, Andrews PW: High-Content Screening for Chemical Modulators of Embryonal Carcinoma Cell Differentiation and Survival. J Biomol Screen 2011
18. Knight A: Systematic reviews of animal experiments demonstrate poor human clinical and toxicological utility. Altern Lab Anim 2007; 35:641-659
19. Adler S, Lindqvist J, Uddenberg K, Hyllner J, Strehl R: Testing potential developmental toxicants with a cytotoxicity assay based on human embryonic stem cells. Altern Lab Anim 2008; 36:129-140
20. Nau H: Teratogenicity of isotretinoin revisited: species variation and the role of all-trans-retinoic acid. J Am Acad Dermatol 2001; 45:S183-187
21. Murry CE, Keller G: Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell 2008; 132:661-680
22. Sartipy P, Bjorquist P: Concise review: Human pluripotent stem cell-based models for cardiac and hepatic toxicity assessment. Stem cells 2011; 29:744-748
23. Ebert AD, Yu J, Rose FF, Jr., Mattis VB, Lorson CL, Thomson JA, et al.: Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature 2009; 457:277-280
24. Itzhaki I, Maizels L, Huber I, Zwi-Dantsis L, Caspi O, Winterstern A, et al.: Modelling the long QT syndrome with induced pluripotent stem cells. Nature 2011; 471:225-229
25. Moretti A, Bellin M, Welling A, Jung CB, Lam JT, Bott- Flugel L, et al.: Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N Engl J Med 2010; 363:1397-1409 2
6. Lee G, Papapetrou EP, Kim H, Chambers SM, Tomishima MJ, Fasano CA, et al.: Modelling pathogenesis and treatment of familial dysautonomia using patientspecific iPSCs. Nature 2009; 461:402-406
27. Damoiseaux R, Sherman SP, Alva JA, Peterson C, Pyle AD: Integrated chemical genomics reveals modifiers of survival in human embryonic stem cells. Stem cells 2009; 27:533-542
28. Xu Y, Zhu X, Hahm HS, Wei W, Hao E, Hayek A, et al.: Revealing a core signaling regulatory mechanism for pluripotent stem cell survival and self-renewal by small molecules. Proc Natl Acad Sci U S A 2010; 107:8129- 8134
29. Andrews PD, Becroft M, Aspegren A, Gilmour J, James MJ, McRae S, et al.: High-content screening of feederfree human embryonic stem cells to identify prosurvival small molecules. Biochem J 2010; 432:21-33
30. Takahashi T, Lord B, Schulze PC, Fryer RM, Sarang SS, Gullans SR, et al.: Ascorbic acid enhances differentiation of embryonic stem cells into cardiac myocytes. Circulation 2003; 107:1912-1916
31. Wu X, Ding S, Ding Q, Gray NS, Schultz PG: Small molecules that induce cardiomyogenesis in embryonic stem cells. J Am Chem Soc 2004; 126:1590-1591
32. Chen S, Borowiak M, Fox JL, Maehr R, Osafune K, Davidow L, et al.: A small molecule that directs differentiation of human ESCs into the pancreatic lineage. Nat Chem Biol 2009; 5:258-265
33. Ding S, Wu TY, Brinker A, Peters EC, Hur W, Gray NS, et al.: Synthetic small molecules that control stem cell fate. Proc Natl Acad Sci U S A 2003; 100:7632-7637