Addressing kinetic applications in High Content Screening
Posted: 29 September 2008 | Edward Ainscow, Research Scientist, Advanced Science and Technology Laboratory, AstraZeneca and Neil Carragher, Associate Research Scientist, Advanced Science and Technology Laboratory, AstraZeneca | No comments yet
Traditional drug discovery screening assays tend to employ simplistic endpoint assays that often monitor the activity of a single target. While these approaches are amenable to high-throughput screening they provide limited information on how candidate drugs influence complex biological systems that exist in vivo. Such limitations are a contributing factor to high attrition rates of drugs as a consequence of poor efficacy in clinical trials.
Traditional drug discovery screening assays tend to employ simplistic endpoint assays that often monitor the activity of a single target. While these approaches are amenable to high-throughput screening they provide limited information on how candidate drugs influence complex biological systems that exist in vivo. Such limitations are a contributing factor to high attrition rates of drugs as a consequence of poor efficacy in clinical trials.
Traditional drug discovery screening assays tend to employ simplistic endpoint assays that often monitor the activity of a single target. While these approaches are amenable to high-throughput screening they provide limited information on how candidate drugs influence complex biological systems that exist in vivo. Such limitations are a contributing factor to high attrition rates of drugs as a consequence of poor efficacy in clinical trials.
To address these issues, pharmaceutical companies are investing in the development and application of high content screens. Such assays incorporating multiple fluorescent probes, multi-parametric image analysis and advanced statistics offer the potential of readouts which more closely reflect in vivo activities and give early identification of off-target effects. Recently, High Content Screens have been used to profile the mechanism-of-action of small molecule1,2 and biological libraries3. This approach has also been applied to more functional assays such as the scratch wound cell migration assay4.
While such fixed endpoint high content approaches provide a wealth of information on pharmacological effect upon a cell phenotype, a significant limitation of most applications thus far is that the assays catch a single snapshot in time following compound addition. Extension of these approaches to kinetic live-cell studies shall enable a more comprehensive temporal evaluation of phenotypic response and detailed analysis of dynamic cellular events including cell activation, migration, cell division, endocytosis and subcellular translocation.
Recent examples demonstrate how live-cell imaging can provide quantative measurements of dynamic cellular events such as, intracellular calcium flux5, oscillations in NF-KB translocation to the nucleus6 and turnover of integrin-associated focal adhesion structures7. Quantification of such dynamic processes are not possible with high content endpoint assays.
Applications on Fluorescent Imagers
The first generation of high content imaging platforms use automated fluorescence optics to obtain high resolution images of cells in multiwell plates. These imaging platforms such as the Cellomics Arrayscan, InCell Analyser and the MDC ImageXpress have been widely used and deployed across early drug development. The principal use for these instruments tends to be fixed end-point assays. Nevertheless, the majority of platforms do offer a time series acquisition option.
In order for a system to be suitable for kinetic cell-based imaging, there needs to be a number of technical requirements. The most obvious is that the environment that the cells are maintained within is capable of sustaining cellular function. For short term applications, up to 1 hr, this may simply be heating the system to 37°C, and for longer-term assays, there is the additional need to control humidity and CO2 concentration. The platform needs to have stable illumination, stage positioning and objective control with minimal thermal fluctuations. Liquid handling capabilities are essential where an acute response is to be monitored. The kinetic imaging capability can be used as a short-cut during assay optimisation e.g. monitoring the optimal timepoint for fixed endpoint screening of a translocation event. However, kinetic imaging can allow innovative approaches to be taken. An example application which is particularly well suited for the fluorescent imaging platforms is the monitoring of calcium flux. Standard plate readers, such as the FLIPR, have the advantage over the imaging platforms in terms of sensitivity, but they are hampered by the fact that their readout is obtained from a population of cells. As such they are not suitable for measuring oscillations of individual cells or the heterogeneity of response in mixed cell cultures.
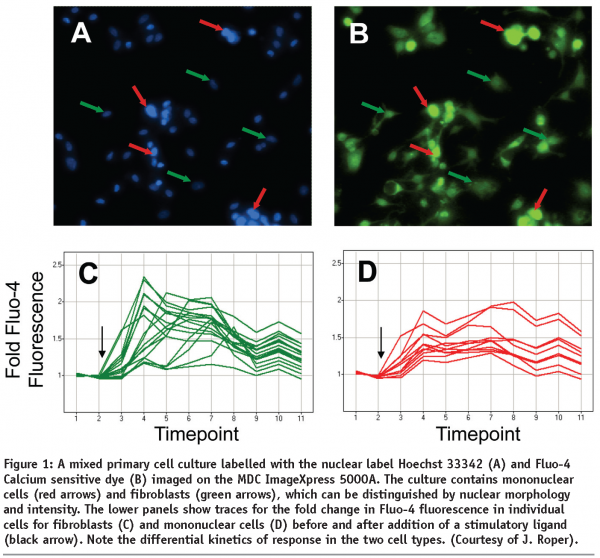

Figure 1 shows the sample results obtained from monitoring GPCR dependent calcium influx in a primary co-culture of fibroblasts and mononuclear cells. Cells were loaded with the calcium sensitive dye, FLUO-4 and the nuclear marker Hoechst 33342 and imaged on an ImageXpresss 5000A system. Distinction can be made between the two cell types based on cellular morphology, with the mononuclear cells having smaller and brighter labelled nuclei than fibroblasts. Note that the mononuclear cells represent ~10% of the population, and as such their response would be outweighed by that of the fibroblasts on a plate reader. Fluorescence intensity of individual cells is monitored pre- and post-application of the ligand, and the kinetics of the response calculated (normalised to the unstimulated fluorescence). By doing so the differential responses of the two cell populations can be characterised. In this case the mononuclear cells display a more sustained response, with a slower time-to-peak, than the fibroblasts.
Advances in Kinetic Imaging Instrumentation
As indicated above, a key consideration in the application of kinetic imaging approaches is to maintain cellular function. As with the platforms above, the configuration is commonly that of a microscope with an environmental chamber attached. An alternative approach is to put the microscope inside an incubator. Such systems are exemplified by the IncuCyteTM and Cell-IQTM. These are automated brightfield and fluorescent imaging microscopes specifically designed for long-term live-cell kinetic studies and represent a significant technological advance in dynamic cell imaging.
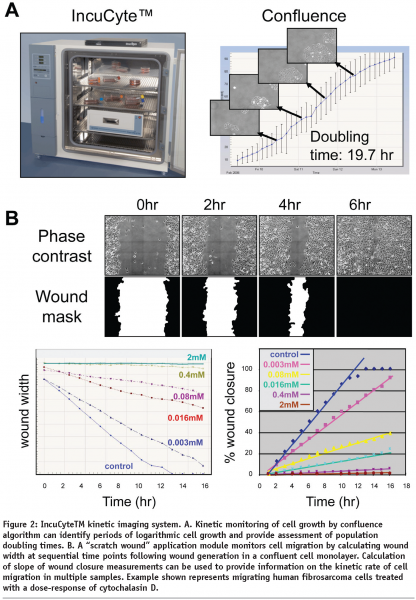
The IncuCyteTM represents a compact, automated, phase contrast imaging system that that can be controlled via remote internet access to acquire sequential images of live cells in multiwell plates or flasks. The IncuCyteTM instrument is designed to be located within a standard laboratory incubator, thereby, ensuring environmental conditions are optimal for long term live-cell studies. The development of a wound healing application module that incorporates a specifically designed tool (WoundmakerTM) to make a uniform wound in a confluent monolayer of cells in each well of a multiwell plate (ImagelockTM) permits automated acquisition and kinetic analysis of cell migration. The IncuCyteTM also provides custom image analysis software that calculates image metrics over time such as “monolayer confluence” and “scratch wound width” or “wound-confluence”. These kinetic measurements enable approximation of cell doubling times and relative velocity of cell migration to be monitored (see Figure 2). Additional detailed kinetic analysis of dynamic changes in cell morphology by third-party software may reveal further mechanistic insight following molecular or pharmacological intervention. Further developments of faster, higher throughput and fluorescent versions of the IncuCyte (IncuCyteHD and IncuCyteFLR) shall increase the throughput and functionality of the IncuCyte system.

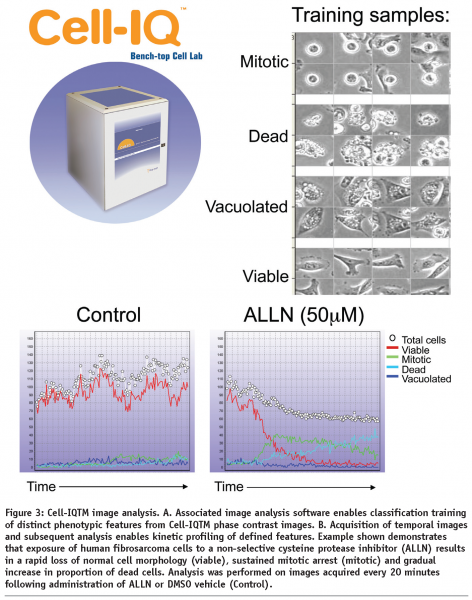
The Cell-IQTM system represents a tissue culture incubator integrated with, phase contrast and fluorescent image acquisition and intelligent analysis software optimized for live-cell studies. The Cell-IQTM instrument enables flexible manual control over field of view and depth of focus and has successfully been used to monitor cell responses in 2-dimensional and 3-dimensional cell culture models (N Carragher unpublished). The Cell-IQ can monitor multiple fields of view from each well of a multiwell plate. Image analysis software associated with the Cell IQTM system utilizes an artificial intelligence training approach to provide robust multiparametric analysis of kinetic live-cell responses without the need for labels or dyes (see Figure 3). However, use of an optional Cell-IQ fluorescent module allows incorporation of fluorescent reporters allowing kinetic analysis of target activity, signalling reporters and phenotypic markers together with label-free metrics.

The Biostation CTTM from Nikon Instruments Inc. represents a more recent development of a phase contrast and fluorescent imaging system for live-cells fully integrated with automated multiwell plate and tissue culture flask handling robotics all within a temperature, CO2 and humidity controlled cell culture incubator.
The development of automated kinetic imaging systems, such as those described above, offer the potential to significantly increase the throughput and scope of live-cell studies by enabling more extensive and robust target identification, validation and secondary screening exercises to be applied to dynamic cell processes. The kinetic profiling of candidate drug responses shall also facilitate the design and application of sequential compound combination and drug wash-out experiments. Quantitative long-term kinetic analysis will also facilitate stem cell differentiation experiments. These approaches can also provide accurate definition of the most appropriate time points for fixed high content assays.
Incorporation of fluorescent reporters into live cell studies allows examination and quantification of specific molecular events. An advance in molecular biology, chemical synthesis and materials sciences has promoted the development of novel fluorescent tools for dynamic imaging of biochemical events in live cells and organisms extending the application of these approaches8. Molecular genetic mutagenesis of natural fluorescent proteins such as green fluorescent protein (GFP) has produced a variety of fluorescent proteins with distinct spectral characteristics that facilitates multiplex analysis of multiple targets or signalling events simultaneously9. The development of, cell permeable, activatable fluorescent probes that monitor key biochemical events such as apoptosis10 or specific enzyme activity such as proteolytic activity11,12 enables kinetic analysis of specific biochemical events or target activity directly in live specimens. Advances in materials science, has also resulted in the development of new classes of synthetic fluorophores such as Quantum dots, which are inorganic nanocrystals composed of a semi-conductor core and ZnS shell that possess discrete fluorescent emission properties13. Quantum dots are characterised by high extinction coefficient and quantum yields that make them brighter and more photostable than fluorescent proteins, properties which are ideal for repeated imaging in live-cell studies.
Emerging techniques for monitoring biochemical events dynamically in live cells and compatible with high content analysis include bimolecular fluorescent complementation (BiFC) and Fluorescence resonance energy transfer (FRET). BiFC monitors protein-protein interactions where interacting protein partners each expressing a split fragment of a fluorescent protein. When the protein partners interact, the split protein fragments complement together forming an intact functional fluorescent protein that acts as a reporter for the protein-protein association14. Engineering of cells that express multiple fluorescent protein complementation pairs allow the dynamic profiling of multiple target pathways in live cells15. FRET monitors the transfer of energy from a donor to an acceptor fluorophore following specific excitation of the donor flurorophore, and subsequent analysis of fluorescent emission from the acceptor. This technique can be used to measure protein-protein interactions, conformational changes or proteolytic cleavage events in live cells. FRET biosensors have been designed to monitor, in live cells, the activity of a number of pharmacological targets16,17. FRET and BiFC permit high spatial and temporal study of enzyme kinetics in live cells, allowing detailed analysis of target activity following molecular and pharmacological intervention.
Image analysis
Application of advanced image analysis algorithms to high content analysis data, allows the simultaneous measurement of multiple cellular characteristics that define phenotypic response1,2. The extension of multiparametric image analysis to live cell studies may reveal further mechanistic insights. A number of software packages are now available that permit sophisticated multiparametric image analysis upon multi-dimensional data sets, for example Image J, Imaris, Image-Pro-Plus and Definiens. One aspect of kinetic imaging which is computationally difficult is the tracking of cells or objects across time points where there is significant movement between images, for example where there is cellular migration or division.
Further development of more sophisticated image analysis software tools that define relationships between multiple dimensions, allow rapid image co-registration between time points and enables automated association of image metrics with longitudinal time points shall further benefit the application of high content analysis to dynamic live cell imaging studies.
Future developments
One limitation of many kinetic imaging approaches is the instability of the fluorescent probes used. Photobleaching often renders fluorescent intensity readouts meaningless during repeated imaging. This is exacerbated by the risk of phototoxicity. More sensitive optics and the use of stable probes such as quantum dots may broaden the applicability of fluorescent imaging, but label free approaches such as phase-contrast imaging (Cell-IQ, IncuCyte) are becoming a viable alternative.
Further development of cell culture consumables or instruments that minimize condensation or temperature dependent expansion/contraction of cell-culture vessels shall facilitate kinetic analysis of live-cell cultures. These advances shall be particularly useful for large screening campaigns where liquid handling robotics may be required to move plates from an environmental chamber onto the microscope stage at sequential timepoints.
Through the combination of biological, engineering and computational developments kinetic imaging is increasingly becoming a practical screening method. Interpretation of responses in 3 and 4 dimensions will give additional insights into the behaviour of candidate drugs.
References
- Perlman ZE, Slack MD, Feng Y, Mitchison TJ, Wu LF, Altschuler SJ (2004) Multidimensional drug profiling by automated microscopy. Science 306:1194-1198
- Tanaka M, Bateman R, Rauh D et al (2005) An unbiased cell morphology-based screen for new, biologically active small molecules. Plos Biol 3:e128
- Moffat J, Grueneberg DA, Yang X et al (2006). A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell; 124(6):1283-98.
- Yarrow JC, Perlman ZE, Westwood NJ, Mitchison TJ (2004) A high-throughput cell migration assay using scratch wound healing, a comparison of image-based readout methods. BMC Biotechnol 4:21-30
- Ainscow EK, Rutter GA (2002). Glucose-stimulated oscillations in free cytosolic ATP concentration imaged in single islet beta-cells: evidence for a Ca2+-dependent mechanism. Diabetes; 51 Suppl 1:S162-70
- Nelson DE, Ihekwaba AE, Elliott M et al (2004) Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science; 306(5696):704-8
- Webb DJ, Donais K, Whitmore LA et al (2004) FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat Cell Biol 6:154-161
- Giepmans BN, Adams SR, Ellisman MH, Tsien RY (2006) The fluorescent toolbox for assessing protein location and function. Science 312:217-224
- Shaner NC, Steinbach PA, Tsien RY (2005) A guide to choosing fluorescent proteins. Nat Methods 2:905-909
- Damianovich M, Ziv I, Heyman S et al (2006) ApoSense: a novel technology for functional molecular imaging of cell death in models of acute renal tubular necrosis. Eur J Nucl Med Mol Imaging;33(3):281-91
- Carragher NO, Fonseca BD, Frame MC (2004) Calpain activity is generally elevated during transformation but has oncogene-specific biological functions. Neoplasia 6:53-73
- Bremer C, Bredow S, Mahmood U, Weissleder R, Tung CH (2001) Optical imaging of matrix metalloproteinase-2 activity in tumors: feasibility study in a mouse model. Radiology 221:523-529
- Bruchez MP (2005) Turning all the lights on: quantum dots in cellular assays. Curr Opin Chem Biol 9:533-537
- Magliery TJ, Regan L. (2006) Reassembled GFP: detecting protein-protein interactions and protein expression patterns. Methods Biochem Anal;47:391-405.
- Michnick SW, MacDonald ML, Westwick JK (2006). Chemical genetic strategies to delineate MAP kinase signaling pathways using protein-fragment complementation assays (PCA). Methods; 40(3):287-93
- Ting AY, Kain KH, Klemke RL, Tsien RY (2001) Genetically encoded fluorescent reporters of protein tyrosine kinase activities in living cells. Proc Natl Acad Sci USA 98:15003-15008
- Vanderklish PW, Krushel LA, Holst BH, Gally JA, Crossin KL, Edelman GM (2000) Marking synaptic activity in dendritic spines with a calpain substrate exhibiting fluorescence resonance energy transfer. Proc Natl Acad Sci USA 97:2253-2258
- Webb DJ, Donais K, Whitmore LA et al (2004) FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat Cell Biol 6:154-161




