Mass spectrometry based proteomics: trends in tools and strategies
Posted: 7 April 2008 | Connie R. Jimenez, OncoProteomics Laboratory, VUmc Cancer Center, Amsterdam | No comments yet
Recent years have seen great upward leaps in the development of mass spectrometry applied to the field of proteomics. Today it is possible to take a complex biological sample such as organelles, cells, tissue or a biofluid, perturbed or stimulated in some way, and identify and quantitate up to several thousand proteins and determine the level of relative change caused by the perturbation or stimulus. The current challenge is not to identify or quantitate proteins in a limited set of samples, but to profile large series (clinical samples, time-course, sub-cellular compartments) at sufficient depth, and to interpret and make biological sense of the data.
Recent years have seen great upward leaps in the development of mass spectrometry applied to the field of proteomics. Today it is possible to take a complex biological sample such as organelles, cells, tissue or a biofluid, perturbed or stimulated in some way, and identify and quantitate up to several thousand proteins and determine the level of relative change caused by the perturbation or stimulus. The current challenge is not to identify or quantitate proteins in a limited set of samples, but to profile large series (clinical samples, time-course, sub-cellular compartments) at sufficient depth, and to interpret and make biological sense of the data.
Recent years have seen great upward leaps in the development of mass spectrometry applied to the field of proteomics. Today it is possible to take a complex biological sample such as organelles, cells, tissue or a biofluid, perturbed or stimulated in some way, and identify and quantitate up to several thousand proteins and determine the level of relative change caused by the perturbation or stimulus. The current challenge is not to identify or quantitate proteins in a limited set of samples, but to profile large series (clinical samples, time-course, sub-cellular compartments) at sufficient depth, and to interpret and make biological sense of the data.
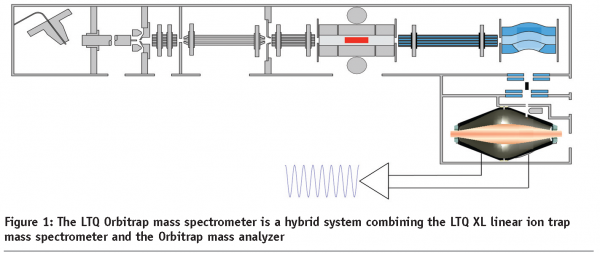
At the end of the 1980s, we saw the birth of two new ionization techniques that changed the face of mass spectrometry; electrospray ionization (ESI) and matrix-assisted laser desorption ionization (MALDI). In the 2000s we have seen terrific developments in mass spectrometer design, such as the development of hybrid fourier-transform, the MALDI-TOF/TOF and hybrid triple quadrupole-iontrap (QTrap) mass spectrometers, and recently an entirely new mass analyser, the orbital electrostatic trap that became incorporated in a hybrid platform with an ion trap (Orbitrap)1 (Figure 1). This new instrument allows for great sensitivity, mass accuracy and high-throughput MS/MS. Combined with the new fragmentation technique ‘Electron Transfer Dissociation’, that preserves labile PTMs great advances are expected in the field of signaling research.
On the wet chemistry side, new approaches have emerged that enable accurate peptide quantitation, such as stable isotope labeling of amino acids in cell culture, isotope tagging reagents for relative and absolute quantitation (iTRAQ), and label-free quantitation using spectral counts and extracted ion abundance. These powerful strategies are rapidly becoming routine forquantifying complex proteomes under different (patho-) physiological conditions. To tackle the complexity of biofluids, especially blood, selective depletion and enrichment strategies are available,enabling profiling of clinical sample series at sufficient depth2. Together with targeted strategies for high-throughput validation of candidate biomarkers, proteomics will deliver its promise to yield new biomarkers for clinical assay development in the coming years. Below I highlight recent research that shows the potential of some of the new proteomics tools.
State-of-the-art in quantitative phosphoproteomics
High throughput quantitative phosphoproteomics is rapidly becoming a reality. Recent state-of-the-art approaches have combined sequential phosphopeptide enrichment (typically strong cat-ion exchange chromatography and immobilised metal-affinity chromatography or titanium oxide), high accuracy identification using LTQ-FTMS or LTQ-Orbitrap and stable isotope labelling. In an impressive recent study3, global, in vivo, site-specific phosphorylation dynamics was obtained for 6.600 phosphorylation sites on 2.244 proteins of Hela cells stimulated with epidermal growth factor (EGF). This study of the dynamic EGF induced phosphoproteome provided a first integrated view of cellular regulation.
Targeted, hypothesis-driven mass spectrometry
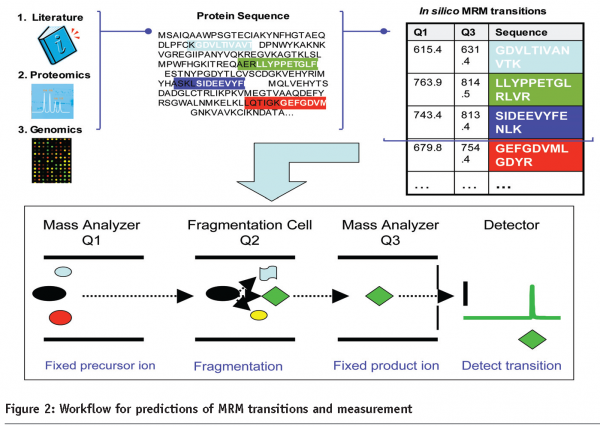
Targeted peptide quantitation is a rapidly growing application within mass spectrometry-based proteomics, due to its potential in biomarker validation, and analysis of signaling pathways through the targeted analysis of post-translational modifications. Selected ion monitoring and more recently multiple reaction monitoring (MRM), techniques widely used in small molecule analysis, have recently been applied to protein analysis4. In the MRM approach, for each protein to be quantified, a monitor (proteotypic) peptide is selected and recorded along with a selected fragment ion (Figure 2). This approach allows for great sensitivity and throughput and when coupled with affinity-based enrichment using new Dynabeads, allows for detection of low abundant candidates5. A promising staged pipeline for generation of valid plasma biomarkers couples, shot-gun sequencing for de novo biomarker discovery to a candidate-based approach to biomarker validation in large sample sets. In a current proof-of-principle study, LC-MS/MS of breast cancer tissue in a mouse model was used for discovery and LC-MRM-MS for validation of biomarker candidates in tissue and blood plasma5.
Another powerful application of LC-MRM-MS is its use for robust quantitation of nodes in signaling networks. In a recent study, stable isotope-labeled peptides were used for highly reproducible MRM-based quantitation of 222 phosphorylation sites across seven time-points following EGF treatment of human mammary epithelial cells6. This targeted strategy enabled the reproducible quantitation of 88% of the peptides, including 31 novel EGF-regulated tyrosine phosphorylation sites.
Towards quantitative organellar proteomics and dynamic cell maps
Biochemical isolation combined with robust mass spectrometry-based proteomics methods has yielded mechanistic insights into the protein composition and function of a diverse range of organelles, including plasma membrane, lipid rafts, nuclear envelope, nuclear pore, synaptic vesicles, post-synaptic density, clathrin-coated vesicles, endosomes, endoplasmatic reticulum, golgi apparatus, mitochondria, peroxisomes, exosomes and more7, To date, quantitation was used to pinpoint organelle-specific proteins, by assessing enrichment of genuine components relative to co-purifying contaminant proteins. In the years to come, the combination of organellar proteomics with perturbations will reveal spatio-temporal information on dynamic protein trafficking between compartments.
Outlook
The new high end hybrid tandem mass spectrometers enable the acquisition of large, consistent datasets across many conditions. For cell biology and signaling research, spatio-temporal data will be indispensable for rigorous computational analysis and mathematical modeling. For biomarker applications, the targeted approaches are expected to bridge the gap between candidate discovery and clinical assays development of validated biomarkers. In short, I foresee a bright future, with mass spectrometry-based quantitative proteomics now turned into a mature field.


References
- Hu Q, Noll RJ, Li H, Makarov A, Hardman M, Graham Cooks R. The Orbitrap: a new mass spectrometer. J Mass Spectrom. 2005 Apr;40(4):430-43.
- Jimenez CR, Piersma S, Pham TV. High-throughput and targeted in-depth mass spectrometry-based approaches for biofluid profiling and biomarker discovery. Biomarkers in Medicine, 2007, in press.
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006 Nov 3;127(3):635-48.
- Anderson, N. L. (2005) The roles of multiple proteomics platforms in a pipeline for new diagnostics. Mol. Cell. Proteomics 4, 1441 –1444
- Whiteaker JR, Zhang H, Zhao L, Wang P, Kelly-Spratt KS, Ivey RG, Piening BD, Feng LC, Kasarda E, Gurley KE, Eng JK, Chodosh LA, Kemp CJ, McIntosh MW, Paulovich AG. Integrated pipeline for mass spectrometry-based discovery and confirmation of biomarkers demonstrated in a mouse model of breast cancer. J Proteome Res. 2007 Oct;6(10):3962-75. Epub 2007 Aug 21.
- Wolf-Yadlin A, Hautaniemi S, Lauffenburger DA, White FM. Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc Natl Acad Sci U S A. 2007 Apr 3;104(14):5860-5. Epub 2007 Mar 26.
- Andersen JS, Mann M. Organellar proteomics: turning inventories into insights. EMBO Rep. 2006 Sep;7(9):874-9.
Connie R. Jimenez
OncoProteomics Laboratory, VUmc Cancer Center, Amsterdam
Connie R. Jimenez, Ph.D. is a biologist with an interest in sub-cellular organelles in relation to function in health and disease. She is head of the recently established OncoProteomics Laboratory in the new Cancer Center Amsterdam research building at the VU University Medical Center. From 1993-1997, during her graduate studies at the Vrije Universiteit in Amsterdam, she pioneered the use of MALDI-TOF mass spectrometry for single cell peptide profiling and for semi-quantitative neuropeptide profiling directly in tissue homogenates. Since her post-doc at UCSF (1997-1999, San Francisco, USA), she has used proteomics as a tool to solve a range of neuroscience-related questions and a current focus is on biomarker discovery in cancer and neurodegenerate disease. In 2000, Dr. Jimenez initiated and is still coordinating the Netherlands Proteomics Platform. Dr. Jimenez participates in several international networks (European Proteomics Association, biomarker discovery committee of the HUPO-Brain project, EU-kp6 consortia cNeuPRO and Angiotargeting and the International Cancer Biomarker Consortium). She is editorial board member of the new Journal of Proteomics.





