Reverse phase protein microarrays for targeted analysis of cellular proteomes
Posted: 13 December 2011 | Ulrike Korf (DKFZ Heidelberg)
In order to advance the identification of new drug targets and disease biomarkers, experimental tools for the systems-level analysis of signalling networks are required. Approaches for a targeted analysis of cellular proteomes have improved in recent years. Notably, the reverse phase protein microarray (RPPA) approach offers great advantages due to properties such as high sensitivity and high sample capacity. This review gives an overview of the principle of RPPA and summarises successful applications that illustrate the potential of RPPA for the analysis of clinical samples, systems biology and for drug discovery concepts. Numerous reports demonstrated the power of this approach to produce higher-order information than is currently possible with any other approach while requiring only minute amounts of sample.
Up-to-date, acquired experience on the application of targeted therapeutics revealed that patients benefit from drugs targeting molecules that are overexpressed by tumours. However, the percentage of patients truly benefiting from the targeted treatment depends largely on the type of tumour. In detail, clinical data obtained from the treatment of solid tumours suggests that our current knowledge is not sufficient to decide beforehand which patients will benefit from a certain treatment and which patients do not. This suggests that overexpression of a particular oncogenic protein by a tumour, such as EGFR, HER2, or oestrogen receptor, does not provide dependable information for treatment decisions. Considerable knowledge has been accumulated on the wiring of those pathways that convey information from cell surface receptors and neighbouring cells as well as the nutritional state and related physiological events. An obvious challenge for proteome research is to convert this knowledge into clinically and pharmaceutically relevant information. However, most drugs target proteins and therefore the realisation of personalised treatment concepts requires a systematic large-scale analysis of individual tumours to identify patterns of deregulation characteristic for subgroups of a certain type of cancer. The identification of reliable disease markers could then be translated into new treatment concepts which have been held back due to technological constraints.
For many years, the analysis of patient-derived samples has presented a technical challenge mostly due to limitations in sample capacity and sensitivity of commonly applied proteomic tools. In general, proteome research has to deal with the fact that protein concentration levels cover several orders of magnitude which embodies high demands for the signal readout dynamics of a certain technology. Due to transcript splicing as well as the fact that the activity of most proteins is regulated by posttranslational modifications (PTM), the number of proteins exceeds the number of genes by at least an order of magnitude. For example, attaching a simple phosphate group to a specific amino acid of a given protein can convert this protein from an inactive to an active state, or vice versa. Thus, proteins function as molecular switches which turn cellular signalling pathways on or off. A systematic proteomic characterisation of individual cancers requires experimental approaches for targeted proteomics to assess the activation state of key players of known signalling networks. A sufficiently high sample capacity to allow the analysis of hundreds of different samples in parallel presents another very important requirement.Reverse phase protein microarray technology (RPPA), introduced a decade ago, has now matured into a promising research tool for the targeted analysis of cellular proteomes.
Reverse phase protein microarrays
The basic idea of RPPA is to assess the abundance of pre-selected target proteins in a large number of samples with target-protein specific antibodies. The method was introduced in 2001 for the quantitative analysis of clinical samples by Emanuel Petricoin and Lance Liotta1. This first report demonstrated that protein abundance in lysates obtained from tumour specimens can be quantified in an arrayed format which is comparable to that of a simple dot-blot procedure. Furthermore, the use of robotic instruments for sample deposition as well as the introduction of suitable sample normalisation measures improved the quantitative readout compared to data obtainable by more subjective methods such as immunohistochemistry.
To date, sample printing relies on high precision robotics and produces an ordered array in which individual samples are addressable by their position on the array. This way, a large number of identical arrays can be produced in parallel so that numerous identical slides result from a single print run. Each slide can be probed with a different target-protein specific antibody (Figure 1). The sample amount delivered per spot usually corresponds to a volume in the low nanolitre range or below. To illustrate this, depositing 1 nanolitre lysate with 2 μg/μL total protein concentration to a solid surface produces a spot containing approximately 2 ng total protein. Thus, with 10 μl lysate as starting material 100 – 200 replicate slides can easily be produced which can subsequently be probed with 100 – 200 different antibodies. In conclusion, RPPA now present a platform that offers high sample capacity so that up to a few thousand samples can be accommodated on a single slide and analysed in parallel.
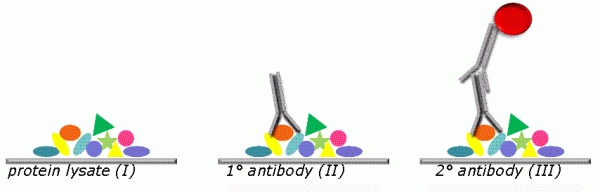
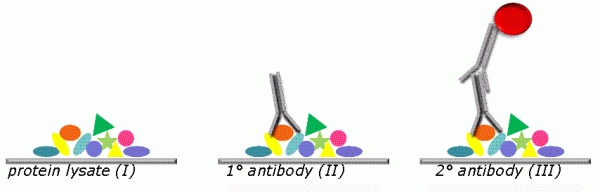
FIGURE 1 Principle of RPPA. Samples are deposited as ordered arrays of droplets. Positive and negative control samples are printed in parallel. Each sample is addressable by its coordinates. The use of highly precise robotic instruments allows printing a large number of identical replicate slides which can be probed with different target-protein specific antibodies. Shown is a single spot (I). Specific protein indicated as orange oval is recognised by target protein-specific antibody (II). Protein/antibody complex is visualised using a secondary antibody which carries a fluorescent dye (III)
It is important to keep in mind that RPPA does not include any protein fractionation step, e.g. by size or charge, and therefore that crude samples are analysed. This circumstance has put a high demand on antibody quality since crossreactivity with other proteins also present in the sample is simply not acceptable. RPPA antibody validation is largely based on Western blot approaches. Usually, cell lysates of suitable controls, e.g. pooled clinical samples or cell lines of interest, are separated by SDS-PAGE and probed by Western blot using a panel of different antibodies directed against proteins of interest. In case of PTM-specific antibodies, cellular signalling might be perturbed by stimulation of specific signalling pathways or induction of apoptosis to enrich those target proteins that are of interest for the analysis. Furthermore, knock-down approaches such as silencing proteins of interest by using siRNAbased approaches turned out to be useful for the validation of antibodies2. Thus, only those antibodies producing highly specific and explainable bands on Western blots in a panel of suitable positive controls are then taken to the next step of antibody validation which is based on probing serial dilutions of lysates by RPPA and correlating RPPA and Western blot signals. However, antibody producers still do not test their antibodies as stringently as required for RPPA and up-to-date characterisation of antibody-specificity needs to be done by the user. Nevertheless, information on RPPA-specific antibodies and robust protocols are becoming publically available3-5 which might help to propagate the application of RPPA further. A modification of the original RPPA approach emphasised the need for sample normalisation measures, and a total protein quantification assay based on the fluorescent dye FAST Green FCF© was introduced. This step allows the calculation of spot-specific correction factors which can be used to normalise the readout obtained from slides incubated with target protein specific antibodies3. Besides that, signal detection relying on near infrared fluorescent dyes resulted in improved signal-to-noise ratios and an increased linear range of signal detection. Significantly improved sensitivity for RPPA was demonstrated by using a detection approach employing the linear waveguide technology for signal detection which eventually resulted in a fully commercialised ‘zeptosens®’ platform for RPPA6.
A quantitative readout of RPPA is based on the fact that a mathematically comprehensible correlation, ideally a sigmoidal curve, is obtained describing the relationship between sample concentration and signal intensity. In principle, the basic idea of all RPPA analysis tools is that only signals within the linear part of the sigmoidal slope are comparable whereas the flanking regions representing signal saturation or the detection limit are excluded from the analysis. RPPA allows analysis of proteins over several orders of magnitude of protein concentration. Software tools7 allow the integration of protein data with experimental or patient-specific information and the resulting data can be visualised as box-plots, heat-maps or time-resolved measurements (Figure 2).
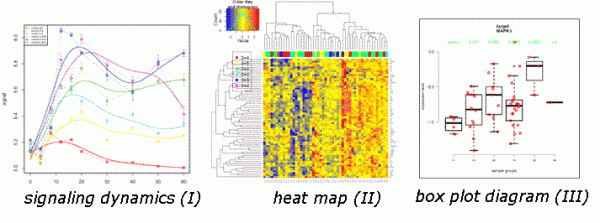

FIGURE 2 RPPA data output. Data can be visualised as box-plot diagrams (I), heat-maps (II) obtained by unsupervised clustering, and as dynamic data obtained in time-resolved measurements (III)
RPPA for personalised medicine in oncology
Worldwide ongoing cancer-genome sequencing efforts aim to accumulate information that might provide a basis for personalised medicine. Clearly, a major challenge of these efforts will be the identification of those mutations that drive the disease, maintain cancerous growth, initiate metastasis and, moreover, that may be relevant for the development of new tailored treatment concepts. In fact, genomic aberrations of cancers affect numerous genes and highly different patterns were identified in individual tumours (e.g. as available via http://cbio.mskcc.org/cancergenomics/gbm/). To translate sequencing information into pharmaceutically relevant information, reverse phase microarrays present the ideal platform to identify signalling pathways preferentially affected by genomic mutations. The protein repertoire of a tumour recapitulates the impact of oncogenic aberrations accumulated during tumorigenesis. In conclusion, analysing the tumour proteome allows us also to study the impact of genomic aberrations. For example, lysates obtained from breast cancer surgical excision specimen were probed for the abundance of 146 target proteins by RPPA. Data identified six molecular subtypes relying on a panel of only 10 protein biomarker profiles which reflect differences in recurrence-free survival8. As a major advantage, RPPA can easily adapt to the clinical routine.
In a technically comparable approach, RPPA-based profiling was carried out for a large number of non small cell lung cancer (NSCLC) cell lines. Data revealed that phosphoproteins sort themselves into pathway-specific activations clusters. Thus, each functional pathway was represented by different signalling proteins. Proteome profiles of NSCLC cell lines with amplified EGFR were highly similar, whereas well-known oncogenic mutations, such as of KRAS for example, executed a comparatively subtle impact on signalling despite the well-described role of oncogeneic KRAS which was exceeded by the impact of effects seen on the phosphoproteomic level (Ummanni et al. manuscript in preparation). In conclusion, the data suggests that mutations of different genes may produce a similar phenotype on the signalling level.
RPPA for the validation of targeted therapeutics
Scientists as well as pharmaceutical companies need to address the question of to what degree individualisation of anti-cancer-treatment concepts can be realised and, moreover, translated into diagnostic products or new drugs. To realise personalised anti-cancer treatment, two requirements need to be fulfilled. First, drug impact on cellular signalling needs to be understood much better on a cellular level and in model systems representative of human cancer subtypes. Secondly, a comprehensive molecular characterisation of cancer diversity is required to identify subtypes that are truly representative for cancer subtypes. Model systems to study drug impact could then be selected based on this knowledge. The current repertoire of RPPAvalidated antibodies covers all cancer-relevant signalling pathways so that time-resolved studies of drug impact can easily identify drug escape mechanisms as shown for the potential of anti-EGFR/ERBB2 directed therapies in breast cancer treatment (Henjes et al. manuscript in preparation).
Reverse phase microarrays to elucidate the role of miRNA
Although the EGFR-driven cell cycle pathway has been extensively studied due to its pivotal role in many different cancers, little is known of how miRNAs coordinate the EGFR protein network on a global miRNA (miRNome) level. Recent reports showed that individual miRNAs can function as a tumour suppressor or oncomirs. Since miRNAs regulate signalling systems by targeting several proteins in parallel, it is not sufficient to analyse the expression or activation of single proteins in response to miRNA overexpression. Again, RPPA also found their place for the analysis of large samples numbers obtained by in vitro experimentation. For example, a large-scale miRNA screening approach using RPPA as readout for targeted proteomics. This way, three miRNAs were identified as novel tumour suppressors that cotarget EGFR-driven cell cycle network proteins and inhibit cell cycle progression and proliferation in cancer (Uhlmann et al. in press).
Summary and outlook
RPPA has matured into a robust experimental platform for targeted proteomics. This platform stands out compared to other tools for targeted proteomics in terms of sensitivity, sample capacity and low sample consumption. Key for a successful application of this platform is access to robust protocols and highly specific antibodies, which is becoming publically available. Thus, in recent years RPPA emerged as a promising tool for targeted proteomics as demonstrated by various successful applications in the fields of drug discovery, miRNA or siRNAlibrary screening as well as for the molecular characterisation of clinical samples and personalised medicine.
References
- Paweletz, C. P., Charboneau, L., Bichsel, V. E., Simone, N. L., Chen, T., Gillespie, J. W., Emmert-Buck, M. R., Roth, M. J., Petricoin, I. E., and Liotta, L. A. (2001) Reverse phase protein microarrays which capture disease progression show activation of prosurvival pathways at the cancer invasion front. Oncogene 20, 1981-1989
- Mannsperger, H. A., Uhlmann, S., Schmidt, C., Wiemann, S., Sahin, O., and Korf, U. RNAi-based validation of antibodies for reverse phase protein arrays. Proteome Sci 8, 69
- Loebke, C., Sueltmann, H., Schmidt, C., Henjes, F., Wiemann, S., Poustka, A., and Korf, U. (2007) Infrared-based protein detection arrays for quantitative proteomics. Proteomics 7, 558-564
- Spurrier, B., Ramalingam, S., and Nishizuka, S. (2008) Reverse-phase protein lysate microarrays for cell signaling analysis. Nat Protoc 3, 1796-1808
- Sevecka, M., Wolf-Yadlin, A., and MacBeath, G. Lysate microarrays enable high-throughput, quantitative investigations of cellular signaling. Mol Cell Proteomics 10, M110 005363
- Pawlak, M., Schick, E., Bopp, M. A., Schneider, M. J., Oroszlan, P., and Ehrat, M. (2002) Zeptosens’ protein microarrays: a novel high performance microarray platform for low abundance protein analysis. Proteomics 2, 383-393
- Mannsperger, H. A., Gade, S., Henjes, F., Beissbarth, T., and Korf, U. RPPanalyzer: Analysis of reversephase protein array data. Bioinformatics 26, 2202- 2203
- Gonzalez-Angulo, A. M., Hennessy, B. T., Meric- Bernstam, F., Sahin, A., Liu, W., Ju, Z., Carey, M. S., Myhre, S., Speers, C., Deng, L., Broaddus, R., Lluch, A., Aparicio, S., Brown, P., Pusztai, L., Symmans, W. F., Alsner, J., Overgaard, J., Borresen-Dale, A. L., Hortobagyi, G. N., Coombes, K. R., and Mills, G. B. Functional proteomics can define prognosis and predict pathologic complete response in




