Forging therapeutics from small interfering RNAs
Posted: 7 March 2005 |
Small interfering RNAs are irreplaceable tools for the functional analysis of pathological gene products. Therapeutic siRNA development leads to new treatment strategies for gene products, where conventional small molecule approaches have failed.
Small interfering RNAs are irreplaceable tools for the functional analysis of pathological gene products. Therapeutic siRNA development leads to new treatment strategies for gene products, where conventional small molecule approaches have failed.
Small interfering RNAs are irreplaceable tools for the functional analysis of pathological gene products. Therapeutic siRNA development leads to new treatment strategies for gene products, where conventional small molecule approaches have failed.
Within four years since their first application in mammalian cell culture, small interfering RNAs (siRNAs) have become essential tools for the analysis of gene function in biomedical sciences. Improvements in chemical RNA synthesis, as well as of delivery systems for siRNAs, has considerably facilitated the application of siRNAs in cell culture. The development of expression cassettes, in combination with the establishment of viral transfection systems for the stable intracellular expression of small hairpin RNAs, provides interesting alternatives to the transient application of preformed siRNAs. Furthermore, the generation of powerful algorithms for the prediction of highly active and specific siRNAs has considerably facilitated siRNA design. As a result of these progresses and due to the high efficiency of such designed siRNAs and the excellent reproducibility of the observed effects, both siRNAs and shRNAs are now widely used for target validation in pharmaceutical chemistry. Moreover, the application of siRNA arrays for the identification of genes involved in cellular processes, such as proliferation, demonstrate the power of RNA interference, not only in the study of a single gene which is implied in a given pathway, but also in the search for new genes not proposed to be involved in a process or pathology so far1.
However, a major fraction of such therapeutically interesting genes cannot be efficiently modulated by, for instance, small molecules with favourable pharmacokinetics. Often, the products of such a gene (RNA or protein) are not ‘drugable’. This leaves the pharmaceutical scientist with the unsatisfying situation that the target is known, but the bullet is missing. Alternatively, one may wish to inhibit only a specific isoform without affecting other isoforms, which may be difficult to achieve with small molecular weight inhibitors. Thus, a bullet is needed but only lead shot is available. In such a situation, it is tempting to think of agents as possible therapeutics that already efficiently and specifically inhibit the gene function in the validation process, but which have unfavourable pharmacological properties in vivo. As already stated, it is comparably easy to design specific and efficient siRNAs. However, their use in therapeutic applications is hampered by low stability in body fluids, very short circulation times in the bloodstream due to rapid renal secretion and inefficient cellular uptake. Despite these limitations, siRNAs are currently making their way through the clinical protocols. This review focuses on the possible therapeutic applications of chemically synthesised siRNAs. The therapeutic application of siRNAs is particularly promising for those diseases where other types of compounds do not exhibit a sufficiently high activity or specificity. For instance, leukaemic fusion genes encoding transcription factors, such as AML1/MTG8, are good examples for potentially valuable therapeutic targets with poor ‘drugability’.
Systemic siRNA delivery
Certain types of therapeutic settings may only require the ex vivo manipulation of cells or tissues with siRNAs. For instance, antileukaemic siRNAs might be used for the purging of haematopoetic stem cells in autologous stem cell transplantation protocols. In such cases, siRNA delivery might be achieved by conventional transfection protocols such as electroporation2. However, the majority of possible applications demand the systemic delivery of therapeutic siRNAs. Currently, the most frequent mode of siRNA delivery in vivo employs the so-called hydrodynamic injection. In this case, a rather large volume of several milliliters of siRNA containing solution is injected within a few seconds into the tail vein of mice. This protocol permits an efficient siRNA delivery particularly to the liver tissue, but is of very limited use for the application in humans. Alternatively, much smaller volumes (200µl or less) of siRNAs may be repeatedly administered by either intravenous or intraperitoneal injection. In each of these protocols, the siRNAs are injected without any transfection agent. The efficacy of, in particular, the intravenous delivery of ‘naked’ siRNAs is limited by poor pharmacokinetics due to rapid renal secretion and to low siRNA stability in body fluids including blood serum3. Nevertheless, several reports describe significant effects in murine disease models upon the delivery of free, unmodified siRNAs4-6. Improved in vivo performance due to chemical modification was shown for siRNA targeting the mouse apolipoprotein B (apoB) gene7. Here, a combination of only three phosphorothioates in the backbone and two 2’-O-methyl groups at ribose sugars gave sufficient protection against exo- and endonuclease degradation. However, this alone was not sufficient to achieve detectable gene silencing in vivo after intravenous injection. In this case, the additional conjugation of a cholesterol moiety to one RNA strand was necessary to deposit sufficient amounts of siRNA in liver and jejunum. Only siRNA harbouring the three types of modifications together, led to inhibition of apoB gene expression in both organs. As a consequence, decreased lipoprotein and cholesterol levels in blood plasma were also achieved. For in vivo applications, increased resistance of siRNA against nucleases were also obtained by using 2’-fluoro8 or 2’-O-methoxyethyl9 modifications of ribose. Locked nucleic acids (LNA) contain a methylene linkage between 2’ and 4’ positions of the ribose. SiRNA containing LNA substitutions possess higher melting temperatures, thus indicating a higher intrinsic stability10. Improved cell delivery of conjugated siRNA was not only observed for cholesterol, but also for lithochloic acid and lauric acid11. Alternative approaches for the systemic siRNA delivery employ transfection agents such as cationic lipids, polyethylenimine (PEI) or its conjugates with polyethylene glycol (PEG) derivatives12-15. All these cationic agents facilitate the endosomal siRNA uptake and the escape of the siRNA from the endosome into the cytoplasm. Particularly the PEI-PEG conjugates show extended circulation times in the blood stream. Those nanoparticles may be modified by the covalent attachment of ligands, such as RGD peptides, to enhance uptake and to specifically target diseased tissues13. Recently, two groups described the efficient intratumourally delivery of siRNA using atelocollagen, a type I collagen derived from calf dermis16,17. Collagen-mediated siRNA administration may be a promising route for the local application of siRNAs.
Preclinical siRNA applications
Therapeutic approaches targeting cancer are frequently limited by reaching an insufficient fraction of tumour cells with the therapeutic compound, by the development of resistance against this compound and by a small therapeutic window due to severe side effects in normal, untransformed tissues. Particularly the last two limitations ask for the identification of targets that are essential for tumour maintenance and which are exclusively expressed in cancerous tissues. Oncogenes carrying exonic mutations are such cancer-specific targets. In contrast to oncogenes with mutated promoter/enhancer sequences or to amplified oncogenes, this class of oncogenes encode mRNAs and proteins with different primary structure compared to the nonmutated proto-oncogene products. Prime examples are mutated p53 forms, constitutively active RAS proteins and the large group of fusion genes generated by chromosomal aberrations. For p53 it has been shown that a single point mutation may be sufficient for siRNAs to discriminate between the mutated oncogene and the nonmutated proto-oncogene18. The siRNA-mediated suppression of leukaemic fusion genes such as BCR-ABL or AML1/MTG8 did not only prove their central roles in tumour formation and maintenance, but also showed the power of siRNAs to interfere with leukaemogenesis in a cancer-specific manner19-21. The presence of several variants of a fusion gene may require the development of siRNAs for each fusion site variant, but, nevertheless, for BCR-ABL all common variants could be efficiently targeted by siRNAs22. Thus, particularly fusion gene-specific siRNAs may become an option to complement existing treatment protocols. However, only relatively small patient numbers are associated with a particular fusion gene, thereby complicating the clinical testing of such siRNAs. Ironically, the high sequence specificity of such antileukaemic siRNAs may hamper their introduction into clinical protocols23. Viral oncogenes may also provide cancer-specific targets. More than 90 per cent of all cervical cancers are positive for human papilloma viruses and express the viral oncogenes E6 and E7. Suppression of these exogenous genes resulted in a reduced proliferation and, dependent on the cell line examined, increased apoptosis or senescence24,25. Thus, siRNA-mediated depletion of the E6 and E7 oncogenes may offer new treatment modalities for cervical cancer and related anogenital tumours. Another group of cancer-relevant targets consists of factors that are neither on the protein nor on the transcript level mutated, but which regulate cellular processes such as apoptosis, cell cycle progression or cellular senescence. In contrast to cancer specific viral or mutated oncogenes, these gene products are also expressed in untransformed tissues. Nevertheless, tumour cells frequently turn out to be more susceptible to the modulation of apoptotic or cell cycle-related gene expression than normal cells. For instance, siRNA-mediated depletion of the antiapoptotic protein BCL-2 induced apoptosis in a variety of cancer cell types and, concomitantly, increased the chemosensitivity of these cells26,27. The inhibition of genes involved in cell cycle progression is another option to interfere with tumourigenic proliferation. Other examples for the successful suppression of proliferation-supportive genes are STAT3 leading to apoptosis in astrocytoma cells. STAT3 siRNAs may, therefore, be promising compounds for the therapy of glioblastomas – brain tumours with extremely poor prognosis28. An alternative strategy to targeting cancer cell proliferation is to interfere with physiological processes essential for tumour growth and maintenance. RNA interference has been used to inhibit tumour angiogenesis by targeting genes promoting neoangiogenesis, such as Vascular Endothelial Growth Factor (VEGF), Colony-Stimulating Factor-1 or Sphingosine 1-Phosphate Receptor-1 with siRNAs. Notably, siRNAs did not only diminish the expression of the corresponding target gene in cell culture, but also delayed tumour growth in mouse model upon systemic siRNA delivery4,29,30. Finally, tumours may not only be directly targeted by siRNAs, but RNA interference may also be used to stimulate an antitumourigenic immune response. Inhibition of apoptosis in antigen presenting cells led to enhanced cancer vaccine potency. In this approach, siRNAs targeting the pro-apoptotic genes BAX and BAK were intradermally administered with a DNA vaccine using a gene gun. The siRNA-mediated suppression of BAX and BAK prolonged the survival time of primed dendritic cells and enhanced cancer-specific CD8+ T cell responses. In contrast to a possible cotransfection with an antiapoptotic gene, this transient administration of siRNAs does not bear the danger of supporting oncogenicity. Thus, such combinations of vaccination and siRNA strategies may hold promise for an enhanced potency of DNA cancer vaccines31. Viral diseases are a second major area where the application of therapeutically active siRNAs is under intense consideration. Influenza virus infection is a major public health problem on a global scale. Every year, several millions cases of severe illness are reported with as many as 500,000 deaths worldwide. Vaccination strategies result in limited protection rates against selected influenza strains. The use of antiviral drugs reduces the severity of the course of the disease, but demands application within a very short time window prior or after infection. Antiviral siRNA may become a new line of defense against virulent strains and is needed for high-risk individuals like infants, elderly or pregnant women. Several siRNA directed against highly conserved regions of the segmented RNA genome of human influenza A were effective. Infected mice showed dramatically reduced viral titers in lung lysates, combined with increased survival rates, when the siRNA was administered prior to the infection or shortly after. The siRNA targeted the genes for nucleocapsid protein and components of the viral RNA polymerase. Interestingly, several routes of administration were successful. The RNA was complexed with a cationic lipid or with polyethyleneimine (PEI) and delivered by instillation directly into the lungs. Interestingly, positive results were obtained, albeit less effective ones, after intravenous hydrodynamic or regular injection of complexed siRNA. A third route of administration used plasmid DNA encoding for antiviral short hairpin RNA32,33. Other viral diseases of the respiratory tract are caused by the respiratory syncytial virus (RSV) and parainfluenza virus (PIV). Both virus types possess medical significance and exert high mortality. Again, no reliable vaccine or antiviral drug exists. These virus genomes possess with the gene for the P protein, a subunit of viral RNA polymerase, at least one promising target gene for RNAi-based therapy. RSV or PIV – infected mice were treated with siRNA-lipid complexes, which were administered intranasal before or after viral infection. For both virus species, decreased virus titers and reduced pulmonary pathology were observed34. The work on all three virus species; Influenza A, RSV and PIV, demonstrated that the lung is an attractive organ for therapeutic siRNA applications as several routes of administration lead to detectable antiviral effects. The investigators also tested the possibility of interferon induction, most notably interferon alpha, by siRNA. No increase in interferon levels were observed, making it unlikely that interferon contributed to virus inhibition besides RNA interference32-34.
siRNAs in clinical trials
The first phase I clinical trials for RNAi therapeutics were already announced. Acuity Pharmaceuticals (Philadelphia, USA) and Sirna Therapeutics (Boulder, USA) have independently developed siRNA as a relief for age-related macular degeneration (AMD). AMD patients suffer vision loss due to neovascularization in eye choroid (CNV) and the subsequent leakage of blood from newly built capillaries into retina. Elevated levels of vascular endothelial growth factor (VEGF) are stimulating capillary growth. Whereas Acuity reduces VEGF levels by administering their lead candidate Cand-5 into the eye, Sirna’s best candidate Sirna-027 is capable of reducing VEGF receptor level. Both companies claim to improve disease progress in animal models (www.sirna.com and35).
Outlook
In this review, we have discussed several examples of therapeutically promising siRNAs and possibilities of administrating them to diseased tissues. The breath-taking progress of taking siRNAs into clinics, however, is based on the experience gained with other antisense molecules such as oligonucleotides and ribozymes. In this respect, one should not forget that many problems such as limited specificity and off-target effects now associated with siRNAs are well known for other oligonucleotide-based technologies as well. Nevertheless, siRNAs will probably be used to complement existing treatment protocols or to establish new treatment modalities. For instance, several reports describe the synergistic effects of siRNA-mediated gene suppression and chemotherapy both in cell culture and in animal models6,36,37. Nevertheless, be it by itself or in combination with other therapeutic agents, RNA interference will very likely expand our possibilities to combat human diseases.
Acknowledgements
I thank Matthias John for his major input into this article. Work in the author’s laboratory is supported by the Deutsche Krebshilfe (10-2104-He2), the Deutsche José Carreras Leukämiestiftung (DJCLS-R03/10) and the Wilhelm-Sander-Stiftung (2003.169.1). 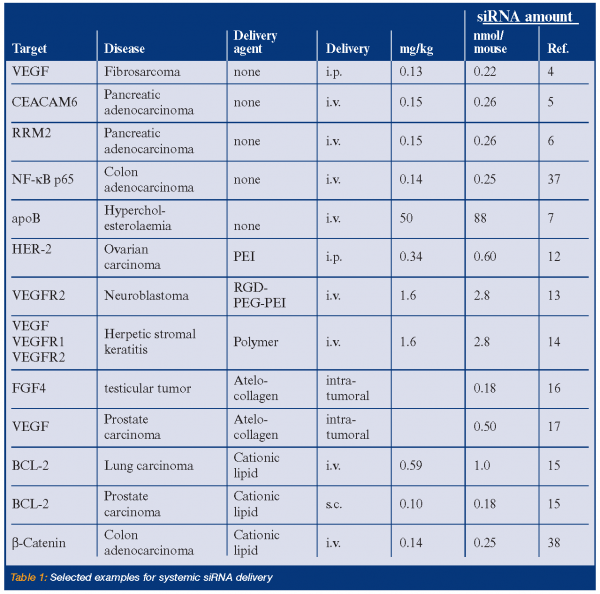

References
- Kittler R, Putz G, Pelletier L, et al. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature. 2004;432:1036-1040.
- Ptasznik A, Nakata Y, Kalota A, Emerson SG, Gewirtz AM. Short interfering RNA (siRNA) targeting the Lyn kinase induces apoptosis in primary, and drug-resistant, BCR-ABL1(+) leukemia cells. Nat Med. 2004;10: 1187-1189.
- Braasch DA, Paroo Z, Constantinescu A, et al. Biodistribution of phosphodiester and phosphorothioate siRNA. Bioorg Med Chem Lett. 2004;14:1139-1143.
- Filleur S, Courtin A, Ait-Si-Ali S, et al. SiRNA-mediated inhibition of vascular endothelial growth factor severely limits tumor resistance to antiangiogenic thrombospondin-1 and slows tumor vascularization and growth. Cancer Res. 2003;63:3919-3922.
- Duxbury MS, Matros E, Ito H, Zinner MJ, Ashley SW, Whang EE. Systemic siRNA-mediated gene silencing: a new approach to targeted therapy of cancer. Ann Surg. 2004;240:667-674; discussion 675-666.
- Duxbury MS, Ito H, Zinner MJ, Ashley SW, Whang EE. RNA interference targeting the M2 subunit of ribonucleotide reductase enhances pancreatic adenocarcinoma chemosensitivity to gemcitabine. Oncogene. 2004;23: 1539-1548.
- Soutschek J, Akinc A, Bramlage B, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173-178.
- Layzer JM, McCaffrey AP, Tanner AK, Huang Z, Kay MA, Sullenger BA. In vivo activity of nuclease-resistant siRNAs. Rna. 2004;10:766-771.
- Dorn G, Patel S, Wotherspoon G, et al. siRNA relieves chronic neuropathic pain. Nucleic Acids Res. 2004;32:e49.
- Braasch DA, Jensen S, Liu Y, et al. RNA interference in mammalian cells by chemically-modified RNA. Biochemistry. 2003;42:7967-7975.
- Lorenz C, Hadwiger P, John M, Vornlocher HP, Unverzagt C. Steroid and lipid conjugates of siRNAs to enhance cellular uptake and gene silencing in liver cells. Bioorg Med Chem Lett. 2004;14:4975-4977.
- Urban-Klein B, Werth S, Abuharbeid S, Czubayko F, Aigner A. RNAi-mediated gene-targeting through systemic application of polyethylenimine (PEI)-complexed siRNA in vivo. Gene Ther. 2004.
- Schiffelers RM, Ansari A, Xu J, et al. Cancer siRNA therapy by tumor selective delivery with ligand-targeted sterically stabilized nanoparticle. Nucleic Acids Res. 2004;32:e149.
- Kim B, Tang Q, Biswas PS, et al. Inhibition of ocular angiogenesis by siRNA targeting vascular endothelial growth factor pathway genes: therapeutic strategy for herpetic stromal keratitis. Am J Pathol. 2004;165:2177-2185.
- Yano J, Hirabayashi K, Nakagawa S, et al. Antitumor activity of small interfering RNA/cationic liposome complex in mouse models of cancer. Clin Cancer Res. 2004;10:7721-7726.
- Minakuchi Y, Takeshita F, Kosaka N, et al. Atelocollagen-mediated synthetic small interfering RNA delivery for effective gene silencing in vitro and in vivo. Nucleic Acids Res. 2004;32:e109.
- Takei Y, Kadomatsu K, Yuzawa Y, Matsuo S, Muramatsu T. A small interfering RNA targeting vascular endothelial growth factor as cancer therapeutics. Cancer Res. 2004;64:3365-3370.
- Martinez LA, Naguibneva I, Lehrmann H, et al. Synthetic small inhibiting RNAs: efficient tools to inactivate oncogenic mutations and restore p53 pathways. Proc Natl Acad Sci U S A. 2002;99:14849-14854.
- Wilda M, Fuchs U, Wossmann W, Borkhardt A. Killing of leukemic cells with a BCR/ABL fusion gene by RNA interference (RNAi). Oncogene. 2002;21:5716-5724.
- Scherr M, Battmer K, Winkler T, Heidenreich O, Ganser A, Eder M. Specific inhibition of bcr-abl gene expression by small interfering RNA. Blood. 2003;101:1566-1569.
- Heidenreich O, Krauter J, Riehle H, et al. AML1/MTG8 oncogene suppression by small interfering RNAs supports myeloid differentiation of t(8;21)-positive leukemic cells. Blood. 2003;101:3157-3163.
- Wohlbold L, van der Kuip H, Moehring A, et al. All common p210 and p190 Bcr-abl variants can be targeted by RNA interference. Leukemia. 2004.
- Borkhardt A, Heidenreich O. RNA interference as a potential tool in the treatment of leukaemia. Expert Opin Biol Ther. 2004;4:1921-1929.
- Jiang M, Milner J. Selective silencing of viral gene expression in HPV-positive human cervical carcinoma cells treated with siRNA, a primer of RNA interference. Oncogene. 2002;21:6041-6048.
- Hall AH, Alexander KA. RNA interference of human papillomavirus type 18 E6 and E7 induces senescence in HeLa cells. J Virol. 2003;77:6066-6069.
- Wacheck V, Losert D, Gunsberg P, et al. Small interfering RNA targeting bcl-2 sensitizes malignant melanoma. Oligonucleotides. 2003;13:393-400.
- Cioca DP, Aoki Y, Kiyosawa K. RNA interference is a functional pathway with therapeutic potential in human myeloid leukemia cell lines. Cancer Gene Ther. 2003;10: 125-133.
- Konnikova L, Kotecki M, Kruger MM, Cochran BH. Knockdown of STAT3 expression by RNAi induces apoptosis in astrocytoma cells. BMC Cancer. 2003;3:23.
- Aharinejad S, Paulus P, Sioud M, et al. Colony-stimulating factor-1 blockade by antisense oligonucleotides and small interfering RNAs suppresses growth of human mammary tumor xenografts in mice. Cancer Res. 2004;64:5378-5384.
- Chae SS, Paik JH, Furneaux H, Hla T. Requirement for sphingosine 1-phosphate receptor-1 in tumor angiogenesis demonstrated by in vivo RNA interference. J Clin Invest. 2004;114:1082-1089.
- Kim TW, Lee JH, He L, et al. Modification of professional antigen-presenting cells with small interfering RNA in vivo to enhance cancer vaccine potency. Cancer Res. 2005;65:309-316.
- Ge Q, Filip L, Bai A, Nguyen T, Eisen HN, Chen J. Inhibition of influenza virus production in virus-infected mice by RNA interference. Proc Natl Acad Sci U S A. 2004;101:8676-8681.
- Tompkins SM, Lo CY, Tumpey TM, Epstein SL. Protection against lethal influenza virus challenge by RNA interference in vivo. Proc Natl Acad Sci U S A. 2004;101:8682-8686.
- Bitko V, Musiyenko A, Shulyayeva O, Barik S. Inhibition of respiratory viruses by nasally administered siRNA. Nat Med. 2005;11:50-55.
- Reich SJ, Fosnot J, Kuroki A, et al. Small interfering RNA (siRNA) targeting VEGF effectively inhibits ocular neovascularization in a mouse model. Mol Vis. 2003; 9:210-216.
- Wohlbold L, Van Der Kuip H, Miething C, et al. Inhibition of bcr-abl gene expression by small interfering RNA sensitizes for imatinib mesylate (STI571). Blood. 2003;102:2236-2239.
- Guo J, Verma UN, Gaynor RB, Frenkel EP, Becerra CR. Enhanced chemosensitivity to irinotecan by RNA interference-mediated down-regulation of the nuclear factor-kappaB p65 subunit. Clin Cancer Res. 2004;10:3333-3341.
- Verma UN, Surabhi RM, Schmaltieg A, Becerra C, Gaynor RB. Small Interfering RNAs Directed against beta-Catenin Inhibit the in Vitro and in Vivo Growth of Colon Cancer Cells. Clin Cancer Res. 2003;9:1291-1300.




