Advances in two-dimensional cell migration assay technologies
Posted: 1 November 2010 |
Cell motility plays an important role in many human diseases and normal cellular processes. Cell migration is critical for wound healing as cells of the inflammatory system and fibroblasts populate the wound and initiate re-epithelialisation1. On the other hand, unregulated cell migration contributes to cancer cell invasion and metastasis2. Agents that affect cell motility, either positively or negatively, could therefore find applications as promoters of wound healing or as antimetastatic drugs. Cell migration in a biological context is an extremely complex process and the understanding of genetic and biochemical determinants remains incomplete.
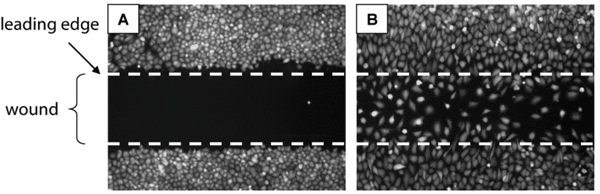

Figure 1 Principle of 2D cell migration assays. (A) A confluent monolayer of cells is mechanically wounded (‘scratch assay’), usually with a sterile pipette tip, leaving two wound edges (dashed lines) separated by a void. Cells at the leading edges quickly assume a polarised morphology and form broad lamellae pointing into the direction of the void. Over time, cells migrate into the void and, eventually, completely close the wound (B). Cell migration is qualitatively assessed by visual inspection and can be quantitated by measuring gap width or by enumeration of cells populating the wound. Images show T98G human glioblastoma cells immediately after wounding (A) or after 24 hours of migration (B). Distance between lines is one millimetre
Cell motility plays an important role in many human diseases and normal cellular processes. Cell migration is critical for wound healing as cells of the inflammatory system and fibroblasts populate the wound and initiate re-epithelialisation1. On the other hand, unregulated cell migration contributes to cancer cell invasion and metastasis2. Agents that affect cell motility, either positively or negatively, could therefore find applications as promoters of wound healing or as antimetastatic drugs. Cell migration in a biological context is an extremely complex process and the understanding of genetic and biochemical determinants remains incomplete.
Contemporary drug discovery paradigms, which rely heavily on single-target, high throughput screening (HTS) fail to capture this complexity. Cell-based assay technologies, in particular image based, high-content screening (HCS)3, provide a promising alternative for the discovery of agents that affect cell motility but have not reached the level of automation required to interrogate large scale compound libraries. The principal barrier to performing HTS of drugs affecting cell migration is a lack of automationcompatible assays that are robust, reproducible and cost-effective to perform. Essentially all existing cell migration assays require mechanical processing steps that are not automatable. In this article, we describe a newly developed cell migration assay that does not involve any mechanical processing steps and present strategies to analyse results by automated image analysis.
Types of cell migration assays
Cell migration assays have been used extensively for more than a decade4,5 but have not made their way into contemporary drug discovery paradigms encompassing large scale screening. Recently, advances in laboratory automation, cellular imaging and compu – tational capacity have revived interest for cell migration assays as evidenced by a number of methods papers in high-profile journals6,7. The most commonly used assay formats are transmigration assays using modified Boyden Chambers with transwell inserts, scratchwounding assays and assays that use mechanical barriers to permit precise timing of cell migration initiation. Transwell assays measure cell migration through porous membranes whereas the latter two detect twodimensional (2D) movement of attached cells on solid surfaces. This article focuses on 2D cell migration assays as they provide the ability to monitor cells in real time during the experiment, are compatible with HCS methodology and have greater potential for HTS implementation.
Scratch wound assay
Probably the most popular assay to study cell migration in vitro is the scratch wounding assay. The scratch assay is performed by creating a cell-free gap, or ‘scratch’, on a confluent cell monolayer upon which cells at the edge of the opening move inward to close the gap (Figure 1). Cell migration is usually assessed by comparing images captured immediately after wounding and at userdefined intervals during scratch closure, generating endpoint-based measurements of wound closure and velocity measurements7,8. The scratch assay is straightforward to perform and inexpensive. However, methods for creating the scratch vary from laboratory to laboratory and results can be highly variable. Furthermore, the process of scratch formation on coated surfaces has been shown to damage the underlying extracellular matrix9. Attempts have been made to implement the scratch assay for HTS. Yarrow et al. used a floating pin tool to inflict a wound to all wells on a 384 well plate10. The resulting wounds could then be analysed with imaging software. While it is possible that this approach could achieve HTS performance, this technique is likely to encounter some reluctance as one could envision damage to both the surface and to the pin tool. Another important consideration of the scratch assay is that cells are injured in the process. This is beneficial in wound healing where growth and survival factors that facilitate wound repair and migration are released locally from injured cells, but it may be undesirable in cancer cell motility assays, which aim to measure abnormal or unregulated mobility in the absence of injury.


Figure 1 Principle of 2D cell migration assays. (A) A confluent monolayer of cells is mechanically wounded (‘scratch assay’), usually with a sterile pipette tip, leaving two wound edges (dashed lines) separated by a void. Cells at the leading edges quickly assume a polarised morphology and form broad lamellae pointing into the direction of the void. Over time, cells migrate into the void and, eventually, completely close the wound (B). Cell migration is qualitatively assessed by visual inspection and can be quantitated by measuring gap width or by enumeration of cells populating the wound. Images show T98G human glioblastoma cells immediately after wounding (A) or after 24 hours of migration (B). Distance between lines is one millimetre
2D migration assays that do not involve cell injury
A gentler version of the scratch wound assay is the Oris™ Cell Migration Assay from Platypus Technologies, LLC (Madison, WI). The Oris™ Assay uses a 96-well plate populated with silicone stoppers that exclude cells from a central detection zone. After cells are seeded and allowed to adhere, the silicone stoppers are removed to reveal an unseeded region in the centre of each well, into which cells are permitted to migrate. Cell migration can be quantified on fluorescence plate readers after labelling cells with live cell dyes such as carboxymethyl fluorescein diacetate (CMFDA) or calcein acetoxymethylester (Calcein AM). To avoid interference from surrounding, non-migrated cells, a mask containing openings similar in diameter to the silicone stoppers (two millimetres) is attached to the bottom of the microplate, limiting the readout to the centre of the well. Alternatively, cell migration can be measured by image analysis. Figure 2 illustrates possible strategies to quantitate migration using images acquired on an ArrayScan HCS reader (ThermoFisher Cellomics, Pittsburgh, PA). A-549 human lung cancer cells labelled with CMFDA were plated in the wells of a 96-well Oris™ plate. After a 24 hour attachment period, a void was created by removing stoppers from all test wells. For pre-migration controls, stoppers remained in wells until assay readout. Cells quickly started to close the gap from the edge; inclusion of hepatocyte growth factor (HGF) accelerated migration (Figure 2A). Migration was first quantified by measurement of the void area by third party software (Definiens Developer, Munich, Germany) (Figure 2B, E), similar to what has been described by other groups using different software packages such as Image J or MetaVue9,10. An algorithm based on the Cellomics SpotDetector Bioapplication also measured void area but misjudged its size when cell densities in the centre were high, i.e., when cells had completely closed the gap (Figure 2C, F). The most robust measurement was obtained when enumerating migrated cells in a single field of view acquired in the centre of the well (Figure 2D, G), even though the shape of the imaging field was different than that of the exclusion zone. This analysis can most likely be further improved by developing image algorithms that count cells in a region of interest defined by the exclusion zone immediately upon removal of stoppers. Since the intro – duction of the Oris™ Assay, several academic and commercial entities have developed barrierbased migration assays6,9,11, including a commercially available assay (CytoSelectTM , Cell Biolabs, Inc.), although none of them are miniaturised. Furthermore, the barrier inherently restricts access to the assay wells and requires a manual removal step, which is incompatible with automation by liquid handling systems.
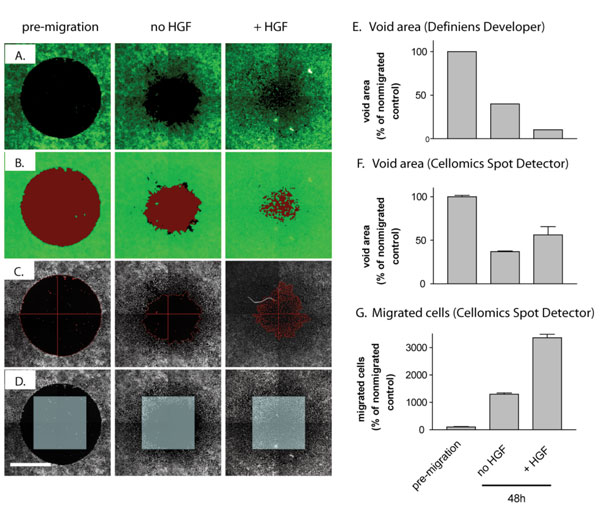

Figure 2 Automated analysis of 2D cell migration in 96-well plates. A-549 cells (40,000 per well) were labelled with CMFDA live cell dye and plated in the wells of a 96- well Oris™ plate. After a 24 hour attachment period, a void area was revealed by removing stoppers from all wells except pre-migration controls. Cells were allowed to migrate for 48 hours in the absence (no HGF) or presence (+ HGF) of 10 ng/ml hepatocyte growth factor, fixed, and nuclei stained with Hoechst 33342. Wells were imaged on an ArrayScan II high-content reader using a 5X objective and a dual (DAPI/FITC) bandpass filter set, and analysed by automated image analysis. A) Raw fluorescence images of CMFDA-labelled cells at the end of the study document cell migration into the void created by the silicone stoppers. B) and E) Void area measurements using Definiens Cognition Network Technology applied to green channel images quantifies void area but requires post acquisition analysis with third party software; green, cell layer, red, void area. C) and F) Void area detection by the Cellomics SpotDetector algorithm permits detection of cells within the gap but misjudges void area when cells have completely closed the gap. D) and G) Enumeration of cells using a single field of view gives the most robust response even though the void shape is different than that of the imaging field. Scale bar, 500 μm
A non-mechanical HTS migration assay
To overcome the disadvantages of barrier-type assays, the Oris™ Assay was modified to replace the silicone stoppers with a chemically engineered cell exclusion zone. The assay, termed Oris™ Pro (Figure 3A) utilises a non-toxic, water-soluble gel spotted into the centre of each well of a 96-well plate, creating uniform areas into which cells cannot penetrate during attachment (Figure 3B). After cells are seeded and allowed to adhere, the gel dissolves to reveal cell-free detection zones in the centre of each well into which cells are permitted to migrate (Figure 3C, D). Because the Oris™ Pro assay does not contain mechanical barriers that impede access to test wells, all assay steps including cell seeding, treatment, and processing are compatible with laboratory automation. The analysis methods described above for HCS are transferable to the Oris™ Pro assay. Figure 4 documents equivalent performance of stopper-based vs. gel-based Oris™ assays using the single field, cell enumeration strategy. MDA-MB231 cells (40,000 per well) were plated in collagen I coated Oris™ or Oris™ Pro plates. After two hours of attachment, stoppers were removed from the Oris™ plate except for eight wells of premigration controls. Medium was removed from both plates and eight wells on the OrisTM Pro plate were fixed and stained with Hoechst 33342 to establish pre-migration controls. The remaining wells on both plates were treated with 10 two-fold concentration gradients of latrunculin A (Sigma, St. Louis, MO, USA) for 48 hours. At the end of the study, stoppers were removed from control wells and cells on both plates were fixed with a formaldehyde solution containing 10 mg/ml Hoechst 33342. A single image field in the centre of each well was acquired on the ArrayScan using a 5x objective and a DAPI compatible filter set and nuclei enumerated. The gel-based assay had a slightly larger assay window and smaller errors compared to the stopper-based assay (Figure 4A). Both assays documented concentration-dependent inhibition of cell migration by latrunculin A (Figure 4B). The concentrations of latrunculin required to elicit half maximal inhibition of cell migration (IC50) were 132 and 135 nM, respectively, for the Oris™ Pro and Oris™ assays.
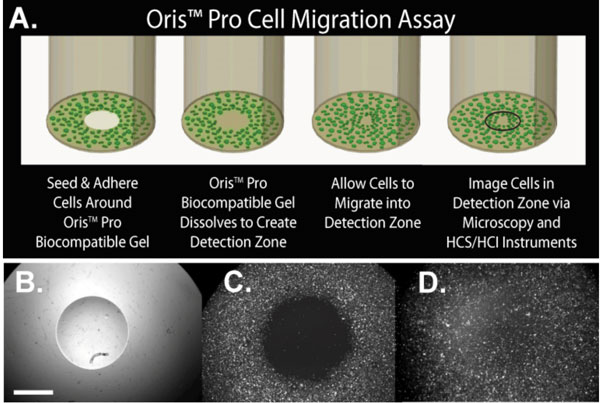

Figure 3 (A) Schematic of the Oris™ Pro assay. (B-D) Temporal presence of cell exclusion zone. Plate is provided with a non-toxic, water soluble gel that creates a cell exclusion zone (B). At four hours post-seeding, CMFDA-labelled HT-1080 cells have attached to a collagen I coated plate at the well perimeter but not in the central exclusion zone (C). At 21 hours post-seeding, cells have migrated to create a monolayer across the entire well (D). All images were taken from the same well. Scale bar, two millimetres Copyright: Images and schematic courtesy of Dr. Keren Hulkower, Platypus Technologies, LLC
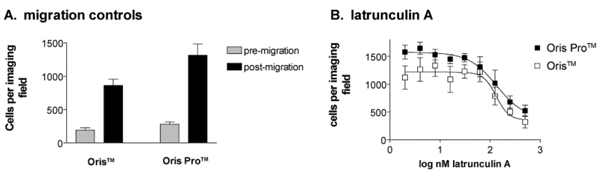

Figure 4 Dose-dependent inhibition of MDA-MB231 cell migration by latrunculin A demonstrates equivalency of Oris™ stopperbased and Oris™ Pro gel-based assays. MDA-MB231 cells were plated in parallel into the wells of collagen I coated Oris™ or Oris™ Pro plates, treated for 48 hours with concentration gradients of latrunculin A, and the number of migrated cells was enumerated on the ArrayScan HCS reader. Both assays measured inhibition of cell migration by latrunculin A in a graded fashion and gave identical IC50 values. Each data point represents the average of four replicates ± S.E
Conclusion and prospectus
Cell motility assays are widely used in drug discovery but their complex setups have limited their application to low throughput secondary or tertiary screens. A promising new alternative is the OrisTM Pro cell migration assay that uses a chemically engineered cell exclusion zone instead of mechanical barriers. The simplified format eliminates mechanical components and renders the assay compatible with HTS instrumentation. Once implemented for 384-well plates, the OrisTM Pro assay will provide a convenient and useful tool to screen large compound libraries for inhibitors or activators of cell migration.
Acknowledgements
I thank Dr. Keren Hulkower, Platypus Tech nologies, LLC, for the images and schematic used to construct Figure 3. This work was supported by U.S. National Institutes of Health grants CA147985 and CA78039.
References
1. Gurtner, G.C., Werner, S., Barrandon, Y., and Longaker, M.T. (2008). Wound repair and regeneration. Nature 453, 314-321
2. Hanahan, D., and Weinberg, R.A. (2000). The hallmarks of cancer. Cell 100, 57-70
3. Giuliano, K.A., Haskins, J.R., and Taylor, D.L. (2003). Advances in high content screening for drug discovery. Assay Drug Dev Technol 1, 565-577
4. Hou, Y.J., Yu, A.C., Garcia, J.M., Aotaki-Keen, A., Lee, Y.L., Eng, L.F., Hjelmeland, L.J., and Menon, V.K. (1995). Astrogliosis in culture. IV. Effects of basic fibroblast growth factor. J Neurosci Res 40, 359-370
5. Savani, R.C., Wang, C., Yang, B., Zhang, S., Kinsella, M.G., Wight, T.N., Stern, R., Nance, D.M., and Turley, E.A. (1995). Migration of bovine aortic smooth muscle cells after wounding injury. The role of hyaluronan and RHAMM. J Clin Invest 95, 1158-1168
6. Kroening, S., and Goppelt-Struebe, M. (2010). Analysis of matrix-dependent cell migration with a barrier migration assay. Sci Signal 3, pl1
7. Liang, C.C., Park, A.Y., and Guan, J.L. (2007). In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc 2, 329-333
8. Vogt, A., Pestell, K.E., Day, B.W., Lazo, J.S., and Wipf, P. (2002). The antisignaling agent SC-alpha alpha delta 9, 4-(benzyl-(2-[(2,5- diphenyloxazole-4-carbonyl) amino]ethyl)carbamoyl)- 2- decanoylaminobutyric acid, is a structurally unique phospholipid analogue with phospholipase C inhibitory activity. Mol Cancer Ther 1, 885-892
9. Kam, Y., Guess, C., Estrada, L., Weidow, B., and Quaranta, V. (2008). A novel circular invasion assay mimics in vivo invasive behavior of cancer cell lines and distinguishes single-cell motility in vitro. BMC Cancer 8, 198
10. Yarrow, J.C., Perlman, Z.E., Westwood, N.J., and Mitchison, T.J. (2004). A high-throughput cell migration assay using scratch wound healing, a comparison of image-based readout methods. BMC Biotechnol 4, 21
11. Poujade, M., Grasland-Mongrain, E., Hertzog, A., Jouanneau, J., Chavrier, P., Ladoux, B., Buguin, A., and Silberzan, P. (2007). Collective migration of an epithelial monolayer in response to a model wound. Proc Natl Acad Sci U S A 104, 15988-15993
About the Author
Andreas Vogt
Andreas Vogt PhD is a Research Assistant Professor in the Department of Pharmacology & Chemical Biology at the University of Pittsburgh and Associate Director of the University of Pittsburgh Drug Discovery Institute. Dr. Vogt has 15 years of experience in small molecule drug discovery. He was Director of Scientific Operations for ProlX Pharmaceuticals (now Oncothyreon, Inc.) and a founding member of the Pittsburgh Molecular Libraries Screening Center (PMLSC). His research interests encompass anticancer drug discovery and high-content screening (HCS) as a discovery tool for challenging drug targets. Dr. Vogt has authored 50 scientific articles and is an investigator on multiple NIH sponsored research projects.
Contact the Author
email: [email protected]




