High Content Analysis approach for targeted gene silencing and probing nanoscale cell responses
Posted: 7 February 2009 | Dr Aideen Long, Senior Lecturer, Trinity College Dublin and Professor Yuri Volkov, Associate Professor of Molecular Medicine, Trinity College Dublin | No comments yet
High Content Analysis and Screening technologies (HCA) ushered in a new era in the biomedical research, enabling the scientists to uncover previously unknown disease mechanisms and to introduce innovative approaches to the development of the new generations of therapeutic drugs with a potential to selectively target individual genes, molecules and subcellular organelles. The quest for the novel therapeutic targets starting from thousands of candidate molecules and compounds, narrowing down to a few leads and their subsequent optimisation and practical application, which for many years has been almost an exclusive privilege of the large pharmaceutical companies, is quickly becoming a routine in the academic research environment.
High Content Analysis and Screening technologies (HCA) ushered in a new era in the biomedical research, enabling the scientists to uncover previously unknown disease mechanisms and to introduce innovative approaches to the development of the new generations of therapeutic drugs with a potential to selectively target individual genes, molecules and subcellular organelles. The quest for the novel therapeutic targets starting from thousands of candidate molecules and compounds, narrowing down to a few leads and their subsequent optimisation and practical application, which for many years has been almost an exclusive privilege of the large pharmaceutical companies, is quickly becoming a routine in the academic research environment.
High Content Analysis and Screening technologies (HCA) ushered in a new era in the biomedical research, enabling the scientists to uncover previously unknown disease mechanisms and to introduce innovative approaches to the development of the new generations of therapeutic drugs with a potential to selectively target individual genes, molecules and subcellular organelles. The quest for the novel therapeutic targets starting from thousands of candidate molecules and compounds, narrowing down to a few leads and their subsequent optimisation and practical application, which for many years has been almost an exclusive privilege of the large pharmaceutical companies, is quickly becoming a routine in the academic research environment.
Investigations into vast collections of small molecule chemicals, genome-wide siRNA libraries and large sets of functionalised nanomaterials for their potential biomedical utilisation, for instance, in addition to being prohibitively time-and labour-consuming by conventional methods, all inevitably generate an extensive output of information difficult for routine analysis and correct interpretation within a confined time frame and restricted resourses. HCA technology is perfectly designed to overcome these limitations, permitting for multiplexed quantitative “on the fly” processing of large massives of data, in addition giving an opportunity of detailed retrospective data analysis thereby providing efficient solutions even within the shortfalls of manpower and funding frequently experienced by academic research centres and which have a potential to become more common in the current economic situation worldwide. We provide here an overview of two extremely beneficial HCA application scenarios currently utilised by the researchers at the Institute of Molecular Medicine, Trinity College Dublin, for tackling extensive massives of experimental data in search of siRNA tools targeting inflammation-associated processes, and for screening the safety and intracellular distribution of nanomaterials with promising biomedical application potential.
Screening T cell migration using an siRNA library
LFA-1-stimulated T cell migration, key to the mechanism of extravasation during the inflammatory process requires the rearrangement of the cellular cytoskeleton and polarisation of the cell. Using High Content Analysis (HCA) to identify changes in cell shape (reflective of changes in migratory capability), we utilised a human siRNA library to screen for novel genes involved in T cell migration.
As part of an effective immune system leukocytes continuously migrate from the circulation across the endothelium into tissue and interchange between the bloodstream and the lymphatics. In doing so they can patrol the body for antigen and enter the sites of inflammation. This process is characterised by initial tethering of the leukocyte to the surface of specialised endothelial cells mediated largely by selectin interaction with their carbohydrate ligands expressed on the endothelial surface1-4. This slows leukocyte flow and cells roll along the surface of the endothelium. Chemokines, secreted by cells within the tissue and immobilised on the endothelial cell surface, and shear stress can activate molecules of the integrin family on the rolling cells, leading to leukocyte arrest and subsequent diapedesis across the endothelium5,6. This process of extravasation is facilitated by polarisation of the cell, which crawls and squeezes between endothelial cells. Leukocytes polarise both in terms of shape and subcellular expression of receptors, adhesion and signaling molecules and cytoskeletal elements7,8. Actin, which is distributed throughout the lymphocyte, concentrates at the leading edge and along with microtubules distribute to the trailing edge or uropod (see Figure 1).
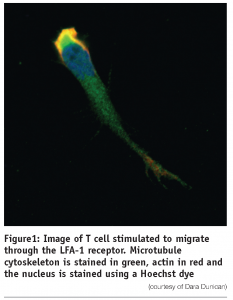

Integrins are a widely expressed family of non-covalently linked α and β subunits that mediate cell-cell and cell-extracellular matrix contacts. The integrins on circulating leukocytes bind minimally to their ligands but their adhesive capacity can be increased by stimulation of intracellular signaling pathways, for example, through the T cell receptor or by chemokines6. This mechanism of activation is termed inside-out signaling. Integrins may also be activated by divalent cations such as Mg2+ and Mn2+ or epitope-specific anti-integrin monoclonal antibodies. Active β 1 and β 2 integrins play an important role in extravasation of leukocytes from the circulation to sites of inflammation and in the homing of lymphocytes to secondary lymphoid organs.
We have developed an in vitro model of T lymphocyte migration9,10. In this system, activated human T lymphocytes (PBTL) or the HuT 78 T cell line migrate (as demonstrated by time-lapse microscopy) when incubated with either immobilised recombinant ICAM-1-Fc fusion protein (the natural ligand for the leukocyte function-associated antigen-1 (LFA-1)) or a triggering antibody to the aL-chain of the LFA-1 molecule itself. T lymphocytes stimulated through the LFA-1 molecule adopt a locomotion-associated polarised phenotype. With this migratory phenotype the cell acquires a long uropod or ‘tail’ which can be several times the actual diameter of the cell body. Upon further analysis of the polarised cells in this model, it was demonstrated that the microtubule and actin components of the cytoskeleton undergo a dramatic rearrangement and is distributed primarily in the trailing uropod of migrating cells (see Figure 1). Expression of the enzyme Protein Kinase C β. which associates with the centrosome and microtubule cytoskeleton in migrating T cells, is necessary for this process of T cell polarisation and migration11.
As T cell migration is central to the inflammatory process, we believe that our model of T cell migration is ideal for use in a screening environment for the study of the mechanism(s) of migration (e.g. the elucidation of signaling/regulatory molecules involved) and for the screening of potential drugs/anti-inflammatory compounds. To this end we have adapted the T cell migration assay to a multi-well format for use in HCA appoach. In this system, culture plates are coated with antibodies or recombinant ligands, which stimulate T cell migration and following polarisation cells are fixed and stained with cytoskeletal markers such as phalloidin (labels actin). The shape of the cytoskeleton is reflective of the migratory capacity of the cell and HCA algorithms such as those which calculate cell elongation or cell area can be used to distinguish round (non-migratory) cells from polarised (migratory) and in some cases hyperpolarised cells. This system is used to screen siRNA libraries and small molecule/ pharmacological compounds to interrogate the mechanisms involved in the migratory process and thus potential pro-inflammatory indicators.
The research group at TCD is currently screening a human siRNA library to identify novel genes involved in the inflammatory process. This genome-wide siRNA library provided by Dharmacon, targets greater than 21,000 genes which are all unique human genes contained in the NCBI RefSeq database. This library is provided in 96-well plates and each gene is targeted by a ‘smartpooltm’ of four siRNA reagents (oligos). The library comprises four sub-libraries:
- The kinome
- G protein-coupled receptor (GPCR)
- Druggable
- Whole genome
The screen may be divided into a number of steps including (i) delivery of siRNA (ii) migration assay (iii) staining of cells (iv) acquisition and analysis of data (v) hit validation/confirmation. These steps are performed in a high throughput environment utilising liquid handling and robotic equipment. Valid hits may subsequently be characterised by various cell/molecular biology methods to elucidate their specific role in the T cell migratory process.
Step 1: delivery of siRNA
Lymphocytes (primary cells or cell lines) are notoriously difficult to transfect, particularly using lipid-based reagents. We use square wave electroporation to deliver the siRNA oligos into these cells (HuT 78 T cell line used for screen). The electroporator (BTX) is equipped with a module which houses a 96-well electroporation plate. Cells are electroporated with siRNA and allowed to ‘rest’ for 72 hours to permit knockdown of protein expression. Typically we observe 80-90% knockdown in those proteins which we have tested.
Step 2: cell migration
As described above, T cells migrate and display a polarised phenotype when stimulated through the β2 integrin receptor, LFA-1. This is achieved by incubating the cells (electroporated with individual siRNA pools) in 96-well plates coated with (a) a goat-anti-mouse immunoglobulin (‘capture’) antibody followed by (b) anti-LFA-1 stimulatory antibody. Cell migration is reflected by a rearrangement of the cytoskeleton, which is assayed as a measure of migration in the screen.
Step 3: staining of the actin cytoskeleton
Cells are stimulated to migrate for a two-hour period following which time they are fixed in 4% paraformaldehyde. The actin cytoskeleton, whose shape reflects the degree of cell polarisation and the migratory capacity of the cell, is stained using Phalloidin and the nucleus using a Hoechst dye. All staining steps are carried out using the Matrix Hydra robot, which is a fully automated computer controlled system, fitted with a 96 well head that can dispense liquids directly into 96 or 384 well plates.
Step 4: image acquisition
This stage of the process is conducted using the INCELL 1000 (GE Healthcare). This High Content Screening system has also been fitted with a robotic plate feeder (Kinedx) and bar code reader with a 90 plate capacity. This instrument is capable of acquiring high resolution microscope images from one to four individual fluorescent dye channels. Once images are captured they are transferred to a high capacity server, where they are stored and subsequently analysed.
Step 5: image and data analysis
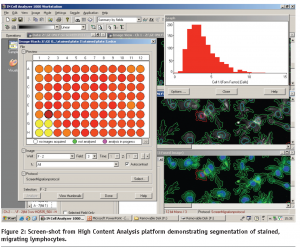
Image analysis involves the use of specialised algorithms, which have been designed to recognise whole cells and/or sub-cellular structures. This HCS image analysis software initially segments individual cells and/or sub-cellular structures (depending on the specific labelling of the cells) (see Figure 2). Once segmentation has been achieved the algorithm can extract information such as staining intensity and morphology parameters such as cell shape and area from these defined cellular/ sub-cellular regions. In the lymphocyte migration siRNA screen, an increase or decrease in cell morphology parameters reflect the effects of each individual siRNA pool on the migratory process. The raw data derived from these cell morphology measurements is then statistically analysed and ranked using the CellHTS software package10.

Step 6: hit validation and characterisation
Genes identified as hits in a siRNA screen must be validated to ensure against off-target effects. A number of strategies or combination of strategies, may be employed to do this including deconvolution of siRNA pools, use of other commercially available siRNAs or use of pharmacological inhibitors of identified hit, if available. In addition, secondary screens may be used which in the case of this screen includes transwell and live cell migration assays. These can be performed in high throughput 96-well format. Hits of particular interest may then be further characterised using cell and molecular biology techniques such as confocal microscopy (including live cell imaging of migrating cells using GFP constructs), overexpression studies or use of constitutively active/dominant-negative constructs.
The screen of the T cell migratory process utilising a human T cell library requires a high throughput, high content approach made possible by a range of technologies. Ultimately, we believe that this approach will identify novel genes involved in lymphocyte migration which may in turn serve as targets for treatment of inflammatory disease.
HCA for characterisation of cell responses to nanomaterials
Rapid development of nanotechnology consistently increases the likelihood of human contact with environmentally presented and manufactured nanomaterials, i.e. the tiny objects ranging in size from one to several hundreds of nanometres and featuring an extreme diversity in shapes and physico-chemical properties (inorganic silica, carbon, metal oxide and inert metal nanoparticles and core/shell structures, polymeric particles and spheres, nanorods, nanotubes, nanowires, dendrimers, liposomes and complex derivatives of all the above to name but a few). However, there is still very little definitive systematic information about the consequences of interactions of nano-scale objects with human cells of diverse origin and therefore safety-related issues are high on the agenda of the recently formulated European Technology Platform on NanoMedicine11. On the other hand, very optimistic expectations are associated with the opportunities of using the nanoparticles as a new class of drug delivery systems, arising from the fact that the finite, but tunable size of the engineered nanostructures used as drug delivery vehicles can impose very precise nano-scale drug distribution barriers at the level of cells, tissues and entire organism thereby eliminating undesirable side effects pertinent to most contemporary medicines. High Content Analysis and Screening approach provide a unique integrated technological toolkit for visualisation and physical characterisation of nanoparticle-cell interactions. An exceptional aspect of this approach is that it is possible to identify individual cell, as well as population responses associated with nanoparticle exposure, as in this way subtle effects on small groups of cells within the whole, which could be averaged out by only screening the whole set, are fully registered and elucidated. We consider that this is a deeply important issue. For example, in nanotoxicology it need not be the typical cell that produces an inappropriate biological response, and disease may emerge from relatively small fractions of the overall population. This issue has never been addressed within the arenas of nanotoxicology, nanomedicine or nanodiagnostics prior to the development of the HCA to its current level.
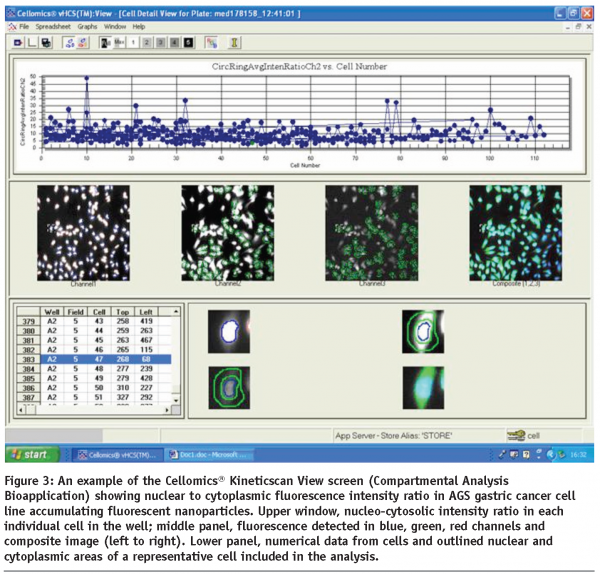
Typical reporter responses of the cells of various origin, including epithelium, fibroblasts, leukocytes and macrophages following the interactions of their membrane surface and receptors with extracellularly delivered nano-objects frequently result in cell spreading, significant shape change and net cell body translocation thereby precluding real-time multi-factorial analysis of reporter events at the cell population level by conventional microscopy methods. The same limitations would apply to the studies implementing expression and dynamic translocation of fluorescently-tagged cytosolic, cytoskeletal and signalling proteins in the sub-membrane compartments of multiple cells interacting with nanomaterials of diverse nature over time. These issues can be effectively eliminated by implementing long-term live cell population scans in HCA devices equipped with controlled microenvironment devices. In addition, HCA technology allows for direct identification of the precise cellular localisation of fluorescence-emitting nanomaterials, either doped with fluorochromes or inherently luminescent particles like quantum dots (see Figure 3) and characterisation of its ability to change dynamically in response to dose-specific treatments.

A number of experimental protocols adopted and tested at the Institute of Molecular Medicine, Trinity College Dublin HCA facility have proven to be suitable for evaluation of multi-parameter cell responses to the nanoparticle exposure. These parameters include nuclear shape and fragmentation, cytoskeletal elements distribution, mitochondrial potential, lysosomal mass and pH, cell permeability, mitotic index, spreading and motility, thereby covering a wide range of physiological, toxic and apoptosis-related cell responses.
The HCA platform is indispensable to other associated cell imaging technologies and provides a powerful tool for such fundamental studies as investigation of key extra- and intracellular factors affecting the specific uptake and intracellular distribution dynamics of the engineered nanoparticles and characterisation of the cell type specificity of the nanoparticle intracellular transport and distribution. Subsequently, the results of analysis of uptake and specific delivery of optimised functionalised nanoparticles can serve as a basis for generation of new potential nanoscale drug delivery systems.
Notably, the HCA-facilitated quest for the nanomaterials suitable for biomedical applications today closely resembles the drug discovery and evaluation process12, however commonly involving an ever increasing amount of interdisciplinary collaborative efforts from physicist, chemists, engineers, and biomedical scientists clearly reflecting the complex nature of the materials under study. It could be summarised in several key technological steps:
Step 1: preparation of stable nanomaterial dispersions suitable for cell culture applications and their primary characterisation
This step has frequently no direct relation yet to the actual medicinal benefits of any particular nanomaterial representing for instance, a putatively “biologically inert” prospective drug carrier (e.g. silicon dioxide nanoparticles). However, it ensures consistency and reproducibility of the subsequent results as well as the possibility to use the future new system in the biologically compatible environment, usually meaning preparation of stable water suspensions. One should keep in mind that determination of even seemingly basic parameters such as size, charge, spectral emission characteristics as well as reliable concentration detection sometimes impose significant initial hurdles in the process.
Step 2: primary screening of candidate nanomaterials
Carried out using the HCA approach in a range of cultured human cell lines, usually selected as representative of the main potential (or expected) exposure routes to the nanomaterials under study, i.e. respiratory, oesophageal, gastric and intestinal epithelial cell lines, hepatic, endothelial, cardiac and phagocytic cells, in the latter case both transformed and primary blood-derived. The assays of choice are usually limited to cell proliferation, nuclear size and segmentation as well as multi-parametric cytotoxicity testing in acute exposure scenario (minutes to several hours). As the outcome of this stage, in addition to the conventional dose response curves generated in the same way as for “conventional” soluble chemical candidates, such parameters as number of particles per cell and exposed cell surface are taken into calculations. Cell seeding density is often an important factor in evaluation of nanoparticle-cell interaction outcomes, since the particle/cell ratios might significantly change in the case of rapid cell population growth. All the assays must be performed at least in triplicates for each experimental point and take into account possible edge effects, to avoid consequent well-to-well volume variability. Independent of the particular plate configuration chosen (96, 384 wells or higher density) it is essential to introduce a maximal degree of speed and automation in cell and reagent handling and distribution, due to the potential issue associated with sedimentation or time-dependent aggregation of some nanomaterials upon contact with cell culture media or cell surfaces.
Step 3: identification of nanomaterials targets and target prioritisation
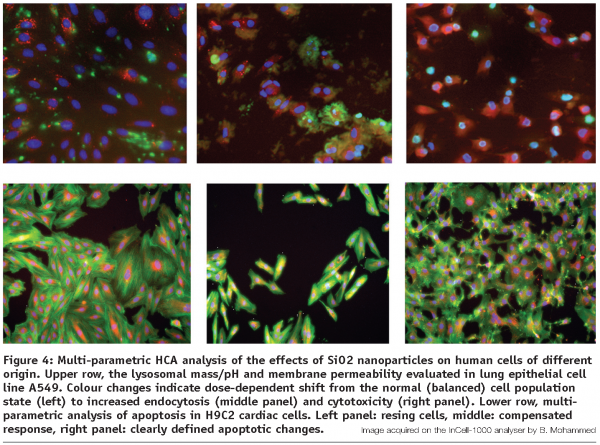
At this stage, following the elimination of the biologically incompatible or acutely toxic candidates identified in step 2, a more detailed, in-depth analysis of cellular uptake, distribution and specific responses to the most promising nanomaterials is being carried out. Same rules as above apply to the general experimental setup, including the handling of the compounds and well-to-well reproducibility. The arsenal of assays commonly includes lysosomal mass/pH evaluation, cytoskeletal elements distribution, cell permeability, cell spreading and motility, multi-parametric apoptosis detection and others, specifically allocated to the particular material under investigation (see Figure 4). Fluorescence-emitting nanoparticles and other labeled (fluorescently doped) nanomaterials can be directly monitored and spatially resolved at their ultimate intracellular locations and specific organelle targets using the HCA devices with confocal capability.

Step 4: identification of leads among the nanomaterials and their optimisation
This process is accomplished through the close feedback interaction of the interdisciplinary research groups via the detailed comparison of the disclosed properties and targeted modification of the identified lead nanomaterials ideally matching particular intended research, diagnostic or drug delivery application and involves repeated smalls scale HCA testing in combination with the full arsenal of all the other complementary physico-chemical and imaging research technologies (spectroscopy, zeta-potential measurements, fluorescence lifetime imaging, live cell confocal microscopy, TEM and others).
In addition to the investigation of nanoparticles as possible carriers in drug delivery or evaluation of their biocompatibility and nanotoxicity, there is a growing demand in the research institutions and pharmaceutical companies worldwide for the availability of multi-colour photo-stable dyes discriminating between cytosolic, nuclear and other subcellular compartments as well as organelle labeling. A significant number of biological assays designed to characterise intracellular transport and functional responses, such as for example, signaling cascades largely exploits molecular translocation events occurring both within the cytoplasm and between cytosol and the nucleus. Contemporary systems of high content analysis, in particular are capable of analysing multiplexed experimental setups implementing a variety of fluorescence-emitting reporters. However, the choice of reliably performing dyes currently on the market is mostly limited to organic dyes with a limited choice of emission wavelengths, e.g. green-emitting calcein and fluorescein derivatives for cytosolic visualisation, blue DAPI and Hoehst range and far-red-emitting DRAQ-5 for nuclear imaging. These either commonly overlap in their emission spectra with a multitude of other popular labels (AlexaFluor, FITC, TRITC etc.) or represent DNA-binding agents emitting in a short-range spectrum (UV) causing significant bleaching of other labels in multi-colour systems and photodamage in live cell studies. A unique opportunity to overcome these limitations is offered by semiconductor quantum dots13-18. As fluorescent probes, quantum dots (QDs) have several advantages over conventional organic dyes, including wide absorption profiles allowing excitation of various QDs of different sizes simultaneously, large linear absorption cross section, narrow emission spectra which are tunable by size and/or composition of QDs, and excellent photostability. Very small (less than 5 nm diameter) negatively charged nanoparticles can accumulate in the cell nuclei (see Figure 5) and display a strong tropism towards the nucleolar histones19.
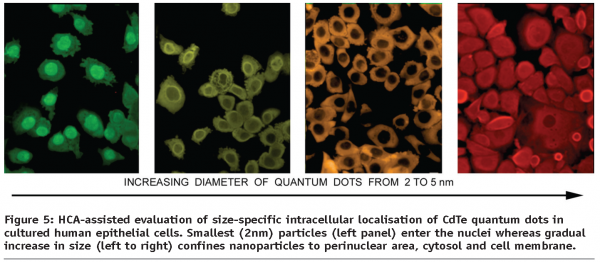

To combat the QDs inherent toxicity, a number of approaches has been suggested for their surface passivation, including the use of PEGs and polymers, rendering modified QDs practically non-toxic in in vitro cell systems and thereby making it feasible to merge the HCA technological approach with new nanoparticle-based fluorescent labels, aiming at delivering a whole class of extremely stable size (and spectrum) separated cell and organelle labels. The experimental studies carried out to date on the intersection of NanoMedicine, BioNanotechnology and HCA has paved the way to establishing the high content screening and analysis approach as a new universal tool for evaluation of the toxicity and specific responses developing in human cells upon exposure to existing and new nanomaterials of diverse origin20.
In conclusion, the high content analysis and screening approach applied to solving of the contemporary biomedical problems enables to reach the results of previously unachievable scientific quality and fundamental significance in diverse areas of research demanding a new level of highly reproducible, quantitative and multi-parametric information.
References
- Chen M, Geng JG. P-selectin mediates adhesion of leukocytes, platelets, and cancer cells in inflammation, thrombosis, and cancer growth and metastasis. Arch Immunol Ther Exp (Warsz). 2006 Mar-Apr;54(2):75-84.
- Ley K, Reutershan J. Leucocyte- endothelial interactions in health and disease. Handb Exp Pharmacol. 2006;(176 Pt 2):97-133.
- Alon R, Dustin ML. Force as a facilitator of integrin conformational changes during leukocyte arrest on blood vessels and antigen-presenting cells. Immunity. 2007 Jan;26(1):17-27.
- Kellersch B, Kolanus W. Membrane-proximal signaling events in beta-2 integrin activation. Results Probl Cell Differ. 2006;43:245-57
- Serrador JM, Vicente-Manzanares M, Calvo J, Barreiro O, Montoya MC, Schwartz-Albiez R, Furthmayr H, Lozano F, Sanchez-Madrid F. A novel serine rich motif in the intercellular adhesion molecule 3 is critical for its ezrin/radixin/moesin-directed subcellular targeting. J Biol Chem. 2002 Mar 22;277(12):10400-9.
- Millan J, Montoya MC, Sancho D, Sanchez-Madrid F, Alonso MA. Lipid rafts mediate biosynthetic transport to the T lymphocyte uropod subdomain and are necessary for uropod integrity and function. Blood. 2002 Feb 1;99(3):978-84.
- Volkov Y, Long A, Kelleher D. Inside the crawling T cell: leukocyte function- associated antigen-1 cross-linking is associated with microtubule-directed translocation of protein kinase C isoenzymes beta(I) and delta. J Immunol. 1998 Dec 15;161(12):6487-95.
- Long A, Mitchell S, Kashanin D, Williams V, Prina Mello A, Shvets I, Kelleher D, Volkov Y. A multidisciplinary approach to the study of T cell migration. Ann N Y Acad Sci. 2004 Dec;1028:313-9.
- Volkov Y, Long A, McGrath S, Ni Eidhin D, Kelleher D. Crucial importance of PKC-beta(I) in LFA-1-mediated locomotion of activated T cells. Nat Immunol. 2001 Jun;2(6):508-14.
- Boutros M, Brás LP, Huber W. Analysis of cell-based RNAi screens. Genome Biol. 2006;7(7):R66.
- http://www.etp-nanomedicine.eu/public
- O’Brien P, Haskins JR. In vitro cytotoxicity assessment.Methods Mol Biol. 2007;356:415-25.
- Xing Y, Rao J. Quantum dot bioconjugates for in vitro diagnostics & in vivo imaging. Cancer Biomark. 2008;4(6):307-19.
- Medintz IL, Mattoussi H, Clapp AR. Potential clinical applications of quantum dots. Int J Nanomedicine. 2008;3(2): 151-67.
- Sukhanova A, Nabiev I. Fluorescent nanocrystal-encoded microbeads for multiplexed cancer imaging and diagnosis. Crit Rev Oncol Hematol. 2008 Oct;68 (1):39-59.
- Biju V, Itoh T, Anas A, Sujith A, Ishikawa M. Semiconductor quantum dots and metal nanoparticles: syntheses, optical properties, and biological applications. Anal Bioanal Chem. 2008 Aug;391(7):2469-95.
- Smith AM, Duan H, Mohs AM, Nie S. Bioconjugated quantum dots for in vivo molecular and cellular imaging. Adv Drug Deliv Rev. 2008 Aug 17;60(11):1226-40.
- Zhang H, Yee D, Wang C. Quantum dots for cancer diagnosis and therapy: biological and clinical perspectives. Nanomed. 2008 Feb;3(1):83-91.
- Conroy J, Byrne SJ, Gun’ko YK, Rakovich YP, Donegan JF, Davies A, Kelleher D, Volkov Y. CdTe nanoparticles display tropism to core histones and histone-rich cell organelles. Small. 2008;4(11):2006-15.
- Jan E., Byrne SJ, Cuddihy M, Davies AM, Volkov Y, Gun’ko YK, Kotov NA. High-Content Screening as a Universal Tool for Fingerprinting of Cytotoxicity of Nanoparticles. ACS Nano 2008;2(5): 928-938.
Acknowledgements
Gabor Bakos, Emily Bennett, Jennifer Conroy, Anthony Davies, Dara Dunican, Antje Hoff, Dermot Kelleher, Bashir Mohammed, Yvonne Williams. Supported by EU Framework 6 (Marie Curie Transfer of Knowledge Programme and NanoInteract Consortium), Health Research Board of Ireland and Science Foundation of Ireland (BioNanoInteract SRC).




