High throughput and high content screening microscopy: A microscopy based guideline
Posted: 30 July 2009 | Jürgen Reymann, Scientist, University of Heidelberg, Holger Erfle, Head of ViroQuant-CellNetworks RNAi Screening Facility, University of Heidelberg, Tautvydas Lisauskas, PhD student, University of Heidelberg and Vytaute· Starkuviene· , Head of the Screening of Cellular Networks research group, BioQuant, University of Heidelberg | No comments yet
The understanding of properties of any biological system requires a detailed and quantitative analysis of its parts and their interactions. As different processes within a system occur at defined space and time, each process holds its own optimal observation and investigation technique. One of the most powerful tools to analyse biological samples quantitatively is based on fluorescence microscopy. Comprehensive studies of diverse biological processes were lately performed by fluorescence screening microscopy, which came up extensively during the last decade1,2,3.
The understanding of properties of any biological system requires a detailed and quantitative analysis of its parts and their interactions. As different processes within a system occur at defined space and time, each process holds its own optimal observation and investigation technique. One of the most powerful tools to analyse biological samples quantitatively is based on fluorescence microscopy. Comprehensive studies of diverse biological processes were lately performed by fluorescence screening microscopy, which came up extensively during the last decade1,2,3.
The understanding of properties of any biological system requires a detailed and quantitative analysis of its parts and their interactions. As different processes within a system occur at defined space and time, each process holds its own optimal observation and investigation technique. One of the most powerful tools to analyse biological samples quantitatively is based on fluorescence microscopy. Comprehensive studies of diverse biological processes were lately performed by fluorescence screening microscopy, which came up extensively during the last decade1,2,3.
This development has strongly been influenced by numerous novel technical improvements like automated focusing and sample positioning, acousto-optical devices for spectral separation and stable light sources which led to the construction of highly automated microscopes. Nevertheless, the different microscope techniques provide their own characteristics and are only suitable for certain biological applications, meaning that the complexity of the application defines the method of data acquisition (DAQ) to be used. The different techniques are optimised in terms of throughput (number of acquired samples per time), high content (e.g., resolution, number of dyes, live cell imaging) and optical quality. A combination of these requirements would thus define the “ideal” microscope for a given application. The main challenge here is to combine high throughput with high content, which unavoidably leads to physical discrepancies due to the optical prerequisites. Furthermore, the classification of biological experiments with regard to the most suitable DAQ does not follow a clear hierarchical structure. In fact, this is defined by several criteria like requirement for resolution, the need for three dimensional (3D) structural resolution and live cell analysis. Another considerable feature is defined by the amount of data to be extracted, regarding the number of biological probes and colour channels to be analysed.
Confocal versus wide field screening microscopy
The mostly used imaging techniques can be related to either focused or non-focused illumination.
Wide field microscopes follow a “standard” optical configuration utilising non-focused illumination. The excitation light is collimated thus illuminating the sample homogeneously over the entire lateral (x-y direction – perpendicular to optical axis) plane. As a result of the homogeneous illumination wide field setups don’t show a well defined depth of field, i.e. resolution in axial (z direction – along optical axis) direction due to the additional detection of out-of-focus light (nevertheless a theoretical value of about 600-1.000 nm using objective lenses with high numerical apertures can be given). Because of their relatively simple optical setup these systems are very robust and rapid in the sense that the entire lateral plane is imaged instantaneously.
In order to obtain real 3D resolution, the elimination of out-of-focus light is indispensable. The excitation light in confocal microscopes is focused at a certain point of the sample and a pinhole in the detection beam path is used which suppresses light from out-of-focus regions from thick specimens (“optical sectioning”)4. In parallel to the gain in information content in the context of 3D resolution, confocal systems lose performance in terms of speed since the focused light beam requires scanning of the sample to image the entire lateral plane. Spinning disk arrangements are able to minimise this limitation but lose slightly in axial resolution instead5.
Wide field configurations: automated wide field microscopes are the preferred systems for high throughput up to genome wide screens (order of magnitude: 105–106 images). Although such microscopes are limited regarding their effective axial resolution and thus structure relevant information, they are very robust and, due to their optical configuration, very rapid compared to point scanning techniques. The minimal requirements for such systems are defined by an arc lamp for excitation of the fluorophores, an automated filter wheel and an automated scanning stage. In addition, the larger part of commercial devices includes adapters to take in standardised multi-well plates or cell arrays of varying formats. Furthermore, the system has to be equipped with specialised screening software for high throughput DAQ. Since the usage of wide field configurations no structural information is needed, such screens generally are run with low resolutions (2x, 10x, 20x or 40x objective lenses). However, data collection utilising objective lenses with high numerical aperture may cause problems due to the lower effective observation volume. In that case, locating of the focal plane for each channel is hindered and, in addition, the effective number of cells to be imaged per region of interest is reduced.
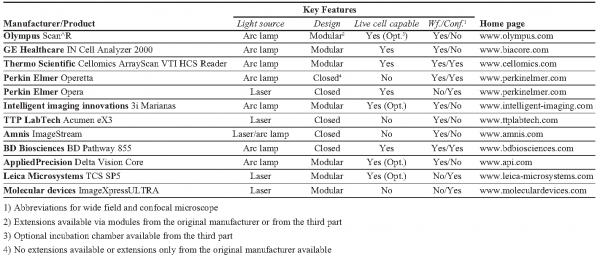
For automated wide field setups the typical DAQ-time of a 384 well plate is about 30 minutes (10x objective lens; 1 position/well; positioning: 500ms; 2 channels; integration time per channel: 200ms; software autofocus: 4s). Utilising a hardware autofocus (1-2 seconds) the overall DAQ-time can be reduced down to 10 minutes (e.g., GE Healthcare IN Cell Analyzer 2000). Following the same optical configuration and using laser excitation, modules based on so called “slide scanning” technologies are available which reach DAQ-times of 10 minutes for every plate type (i.e. up to 1.536 well plates), but with slightly lower imaging quality (e.g., TTP LabTech Acumen eX3).
Confocal configurations: In case the biological question requires 3D structures to be acquired, confocal imaging techniques have to be applied. Confocal microscopes generally utilise laser sources with different wave lengths for excitation of fluorophores and usually enable the simultaneous detection of multiple channels (e.g., Leica TCS SP5, Perkin Elmer Opera). In the case of high resolution, DAQ objective lenses with high numerical aperture (magnification: 40x, 60x/63x and 100x) are used and, in addition to the lateral an axial scanning procedure for acquiring a 3D data stack has to be carried out, DAQ-times are much higher in comparison to wide field configurations. Based on that, a general statement about the time needed for an entire screen is very difficult. A typical value for high-grade automated confocal point scanning devices with hardware autofocus is lying at about one image per second. However, several different factors affect the overall screening time like objective lens, number of slides to be acquired in axial direction, the axial step size and so on. All of these parameters influence the scanning procedure and thus the scanning time essentially. Nevertheless, the use of spinning disk technologies leads to a considerable reduction of the effective DAQ-time in analogy to advanced wide field scanners6. A spinning disk replaces the single objective lens via a special arrangement of micro lenses (as many as 1.000 foci) at a disk which rotates at very high speed (“spinning disk” / “Nipkow disk”). The specific arrangement of the micro lenses enables the scanning of the entire field of view during one rotation cycle. Such systems reach up to 1.000 frames per second (e.g., BD Biosciences BD Pathway 855) and detect up to five channels simultaneously (e.g., Leica TCS SP5). As for advanced wide field scanners the gain in speed comes with a slight loss of imaging quality due to a lower axial resolution compared to point scanning devices5.
Time-critical and non-time-critical screening
For time-critical experiments (e.g., time-lapse experiments, rapid cellular events) the same difficulties hold for every imaging procedure, i.e. for both wide field and confocal techniques. As a rule, time-critical experiments are performed in living cells. There is a set of minimal requirements for performing screens under live cell conditions: live cell imaging means keeping biological samples in a closed environment enabling temperature, humidity and CO2 to be held constant over time. High-grade automated systems generally come with completely closed modules allowing appropriate conditions to be generated (e.g., BD Biosciences BD Pathway 855, Thermo Scientific Array Scan). Furthermore, specifically for the microscope setup of different manufacturers, designed incubator chambers are available. Being able to image living cells, particularly for prolonged times of incubation, means fine temporally resolved information can be obtained7. This allows the ability not only to better identify and classify observed phenotypes, but also to collect data, which could be used to predict properties of a given biological system. Unfortunately, many processes, analysis of which could highly benefit from imaging in living cells, are difficult to establish for a large-scale (e.g., virus cell entry, stimulation of cells with appropriate compounds). For such cases, addition of stimuli and following imaging should be precisely coordinated in time.
Hardly any special requirement for screening systems is needed when doing so called “non-time-critical” experiments. That largely includes screens in fixed cells8,9. Work in living cells could also be considered as “non-time-critical” when no time-lapse acquisition is needed and phenotype under investigation is stably expressed over a longer period of time (e.g., end-point cell viability assays).
Improvement of performance regarding speed, DAQ and resolution
Many biological processes occur within a time frame, which is still difficult to capture in living cells in a large-scale mode. For example, there are number of screens performed to quantify processes of intracellular protein trafficking8,10,11, but virtually all of them are done in fixed cells after selecting the most favourable time point. In order to follow trafficking of cargo proteins or dynamic changes of organelles in living cells, rapid screening microscopy is necessary. In order to achieve this, a number of parameters should be improved or even newly developed.
Parallelisation: Most likely, the easiest and, in relation to effort, the most effective method for reducing DAQ-times is given by a high-grade parallelised excitation and detection of the different colour channels by how the time consuming filter change can be eliminated. For that, the spectral fluorophore characteristics are separated simultaneously either via acousto-optical elements (highly flexible due to continuous and arbitary wave length selection, allowing multiple dye combinations to be used) or pre-defined filter combinations (more limited due to wave length specific colours and thus dye combinations). According to the possible number of channels to be acquired in parallel, multiple sensors (photo multipliers / CCD cameras) attached at the corresponding detection beam path capture the different fluorophore signals in parallel (e.g., Amnis ImageStream). A key role is played by the choice of the fluorophores as well. Both the excitation and the emission spectra of the fluorophores should feature a maximum distance from each other. Otherwise cross talk can be detected which either has to be corrected via image processing or to be avoided by sequential scanning.
Autofocus: Another significant decrease of DAQ-times can be achieved via suitable focusing methods. A software autofocus calculates the maximum intensity measured at the sensor and interprets this as the focus position in generally two to five seconds. But for an effective optimisation, the integration of a hardware autofocus should be chosen which measures changes of the refractive index between dish and solution by using an additional laser source (in general, dark red spectrum (IR) thus avoiding damaging of the probes)12. The reflection pattern is then used to focus at the bottom of the dish within one to two seconds (e.g., Olympus Scan^R). In this context the signal-to-noise ratio (SNR) thus implying high-quality biological samples, probe substrates and so on represents a further class of influences to the overall acquisition due to its direct correlation with exposure time and effort needed during focussing.
Influence of DAQ- and analysis software: Depending on the complexity of the experiment the available software can strongly influence DAQ in terms of time and quality. Most DAQ-programs offer so called focusing maps which can be used within time-laps screens. If more than one imaging cycle of an entire well plate / cell array has to be carried out, the focus positions can be stored for each position and be reused in every following cycle (provided that the focus positions remain constant). Another option is to determine selective focus positions prior to screening and interpolate these values in order to generate a focusing map (e.g., reasonable if large-scale arrays not being subdivided by well borders are used) (e.g., Olympus Scan^R).
In addition, advanced analysis software packages enable online (pre-) processing of the data (e.g., Perkin Elmer Opera, TTP LabTech Acumen eX3). Apart from online process observation, procedures can be realised which, for instance, perform a coarse scanning (e.g., low magnification, 2D image, one time point). If specific pre-defined features of the actual imaged samples are identified via online image processing, an immediate feed-back is sent to the microscope control (DAQ-software) which starts the real scanning procedure (e.g., high magnification, 3D data stack, time-laps) subsequently. This allows, on the one hand, constraining of the data to be acquired to interesting targets, and on the other hand, protection of the samples against bleaching.
High resolution microscopy and screening: standard microscopy techniques as described above underlie the optical diffraction limit (about 180nm in lateral and 500 nm in axial direction (confocal setups) using objective lenses with high numerical aperture (NA = 1.4/1.45)). Modern highest resolution methods like 4Pi13, structured illumination (OMX / SIM)14, STED15 or localisation-based microscopy (PALM / STORM)16 enable structures down to a single molecular level to be resolved thus providing ultra-high information content17. Although such systems are not yet suitable for high throughput procedures, especially localisation-based imaging which in a broader sense can be treated as screening, meaning that a large number of single molecules in cells (instead of a large number of cells in wells) has to be screened. In each case, the resulting data sets contain a number of images in the order of magnitude of 105–106 per channel. Apart from suitable fluorophores, the technical requirements here concentrate as well as on the speed of the screening process in terms of acquiring 2D images, on underlying limiting parameters like correlation between frame rates and detection efficiency.
Conclusion
Nowadays numerous different screening systems capable for high throughput and high content DAQ are available. Nevertheless, the decision concerning the preferred screening system still depends strongly on the biological application to be addressed. Moreover, in addition to specifications listed in official documents, it makes sense to test different screening devices with appropriate biological samples not only in terms of DAQ-times, but also for image quality. In this context, it has to be carefully weighted if the gain in DAQ-times when using plate readers does not lead to non-acceptable compromises in imaging quality. Compared to “real” microscope systems utilising highest quality optical configurations and detectors, plate readers generally show minor image qualities regarding consistency, sensitivity and dynamic range of signal detection18. A further decisive point is related to image processing software. Most of the common manufactures offer software enabling at least standardised analysis of the data (e.g., detection of objects, quantification of fluorescence intensity). Besides that open source image analysis tools are available (e.g., CellProfiler; http://www.cellprofiler.org).
Outlook
Current and future developments concentrate on both a higher flexibility of the optical setup with regard to wide field / confocal configurations, light source, DAQ- / analysis software and a reduction of DAQ-times, although such advanced systems will still not be able to answer numerous fundamental biological questions. However, a completely new research field within the quantitative analysis of biological systems can only be addressed by observing dynamical processes instantaneously over the entire large-scale format of well plates / cell arrays19 containing a high number of diverse samples. First steps into that direction have already been made by highly parallelised screening devices (e.g., d·metrix; http://www.dmetrix.net). Furthermore, intelligent algorithms providing features like unsupervised learning for classification, pattern recognition, particle tracking and so on will help not only to provide interesting results but also to facilitate direct feed-back control to the DAQ-software of the screening microscope during the scanning procedure.

References
- Pepperkok R and Ellenberg J, High-throughput fluorescence microscopy for systems biology, Nat. Rev. Mol. Cell Biol. 7(9):690-696 (2006).
- Paran Y et al., Development and application of automated high-resolution light microscopy for cell-based screens, Meth. Enzymol. 414:228-247 (2006).
- Liebel U et al., A microscope-based screening platform for large-scale functional protein analysis in intact cells, FEBS Lett. 554(3): 394-398 (2003).
- Pawley J, Handbook of Biological Confocal Microscopy, Springer, 3rd Edition (2006).
- Egner A, Andresen V and Hell S, Comparison of the axial resolution of practical Nipkow-disk confocal fluorescence microscopy with that of multifocal multiphoton microscopy: theory and experiment, J. Microsc. 206:24-32 (2002).
- Chong F et al., Optimization of Spinning Disk Confocal Microscopy: Synchronization with the Ultra-Sensitive EMCCD, Proc. SPIE 5324:65-76 (2004).
- Neumann B et al., High-throughput RNAi screening by time-laps imaging of live human cells, Nat. Meth. 3(5):385-390 (2006).
- Pelkmans L et al., Genome-wide analysis of human kinases in clathrin- and caveolae/raft-mediated endocytosis, Nature 436(7047):78-86 (2005).
- Brass A et al., Identification of Host Proteins Required for HIV Infection Through a Functional Genomic Screen, Science 319(5865):921-926 (2008).
- Wassmer T et al., A loss-of-function screen reveals SNX5 and SNX6 as potential components of the mammalian retromer, J. Cell Sci. 120(1):45-54 (2006).
- Das Sarma J, Kaplan B, Willemsen D and Koval M, Identification of rab20 as a potential regulator of connexion 43 trafficking, Cell Com. Adhes. 15(1):65-74 (2008).
- Xiaoqiang W et al., Autofocus methods for automated microscopy, Proc. SPIE, 4224: 114-117 (2000).
- Hell S et al., Measurement of 4pi-confocal point spread function proves 75 nm axial resolution, Appl. Phys. Lett. 64(11): 1335-1337 (1994).
- Gustafsson M, Agard D and Sedat J, Sevenfold improvement of axial resolution in 3D widefield microscopy using two objective lenses, Proc. SPIE 2412:62-65 (1995).
- Hell S, Strategy for far-field optical imaging and writing without diffraction limit, Phys. Lett. A., 326(1-2):140-145 (2004).
- Betzig E et al., Imaging Intracellular Fluorescent Proteins at Nanometer Resolution, Science 313(5793): 1642-1645 (2006).
- Huang B, Wang W, Bates M and Zhuang X, Three-Dimensional Super-Resolution Imaging by Stochastic Optical Reconstruction Microscopy, Science 319(5864):810-813 (2008).
- Bushway P, Mercola M and Price J, A Comparative Analysis of Standard Microtiter Plate reading Versus Imaging in Cellular Assays, ASSAY and Drug Dev. Tech. 6(4): 557-567 (2008).
- Erfle H et al., Reverse transfection on cell arrays for high content screening microscopy, Nat. Prot. 2(2):392-399 (2007).




