Machine learning in biopharmaceutical manufacturing
Posted: 14 September 2018 | André C Guerra, Professor Jarka Glassey | No comments yet
The biotechnology industry is expected to increase the production of new biopharmaceuticals.1 Biopharmaceuticals require high-quality standards, high initial investments for approval and introduction into the market as well as continued investment in manufacturing.2,3


In order to achieve profitable and sustainable manufacturing of biopharmaceuticals, bioprocess and bioproduct development must be planned and executed concurrently throughout the production lifecycle.4 Currently, bioprocess development is a critical bottleneck for the successful implementation of innovation obtained during bioproduct development.5 The majority of host-cell screening, initial conditions, material attributes and bioprocess parameter in-depth optimisation, as well as the identification of relationships between critical process parameters (CPP) and critical quality attributes (CQA), happens at an early stage of development during the implementation and execution of design of experiments (DoE) towards the established quality by design (QbD) paradigm.4,6
Process modelling is required in order to achieve the aforementioned goals.7 Product and process development have been largely accelerated by the development of representative models in the past, mainly in the domain of product discovery and development. However, with the drive-force behind Industry 4.0, the ubiquitous application of process models in flexible, autonomous, scalable biotherapeutical drug manufacturing platforms is expected.1,8 For instance, process modelling will be crucial in advanced biotransformations catalysed by in vitro synthetic (enzymatic) biosystems.9
Nevertheless, current biopharmaceutical manufacturing still requires continuous process / product assessment and evaluation, since different sources of variation – such as environmental disturbances, slow process drifts (ie, fouling, cell / inhibitors / activators activity loss) and process disturbances (ie, feedstock quality / impurities, step / grade inputs, manual sampling) – may influence process outcome.4,10 Several factors contribute to the varying yields still observed in large-scale biopharmaceutical manufacturing, such as the difficulty in balancing biosynthesis of pharmaceuticals with the inherent cell physiology conditioned by the operating conditions.11 All bioprocesses exhibit nonlinear, dynamic behaviours to some extent, which depends on unknown reaction kinetics with time-varying parameters. A variety of factors – including a lack of mechanistic understanding of these biochemical kinetic reactions and instrumentation for online measurements (especially for CQAs); the complex interactions between multiple variables with varying degrees of correlation; time-delayed and scale-dependent bioprocess responses – all introduce new challenges for biotechnologists to build trustworthy representations of the process and product state in the form of a process model.4,10,12
A process model is a mathematical representation of the relationships between process operating parameters and process outputs / state or product quality attributes. Models can describe a single process unit operation, ranging from upstream to downstream unit operations, or provide a holistic representation of the whole process.12 Mechanistic models, relying on first / fundamental principles and phenomenological assumptions, are typically parameterised empirically. The applicability of such mathematical process models, typically composed of a system of differential equations, has been demonstrated in the development of improved strategies for DoE, monitoring and control strategies.12,13 Advances and advantages of mechanistic modelling for biopharmaceutical manufacturing have been reported;11–16 however, mechanistic models have several limitations. For example, the lack of necessary model resolution to predict biopharmaceutical CQAs16 and the static representation of dynamic (scale and time-dependent) parameters.15 Currently, mechanistic models are reportedly used in the monitoring and control of bioprocess outputs, based on the feed rate of raw materials,14,16,17 but the controllability of specific CQAs (such as glycosylation of monoclonal antibodies) is yet to be fully evaluated.17
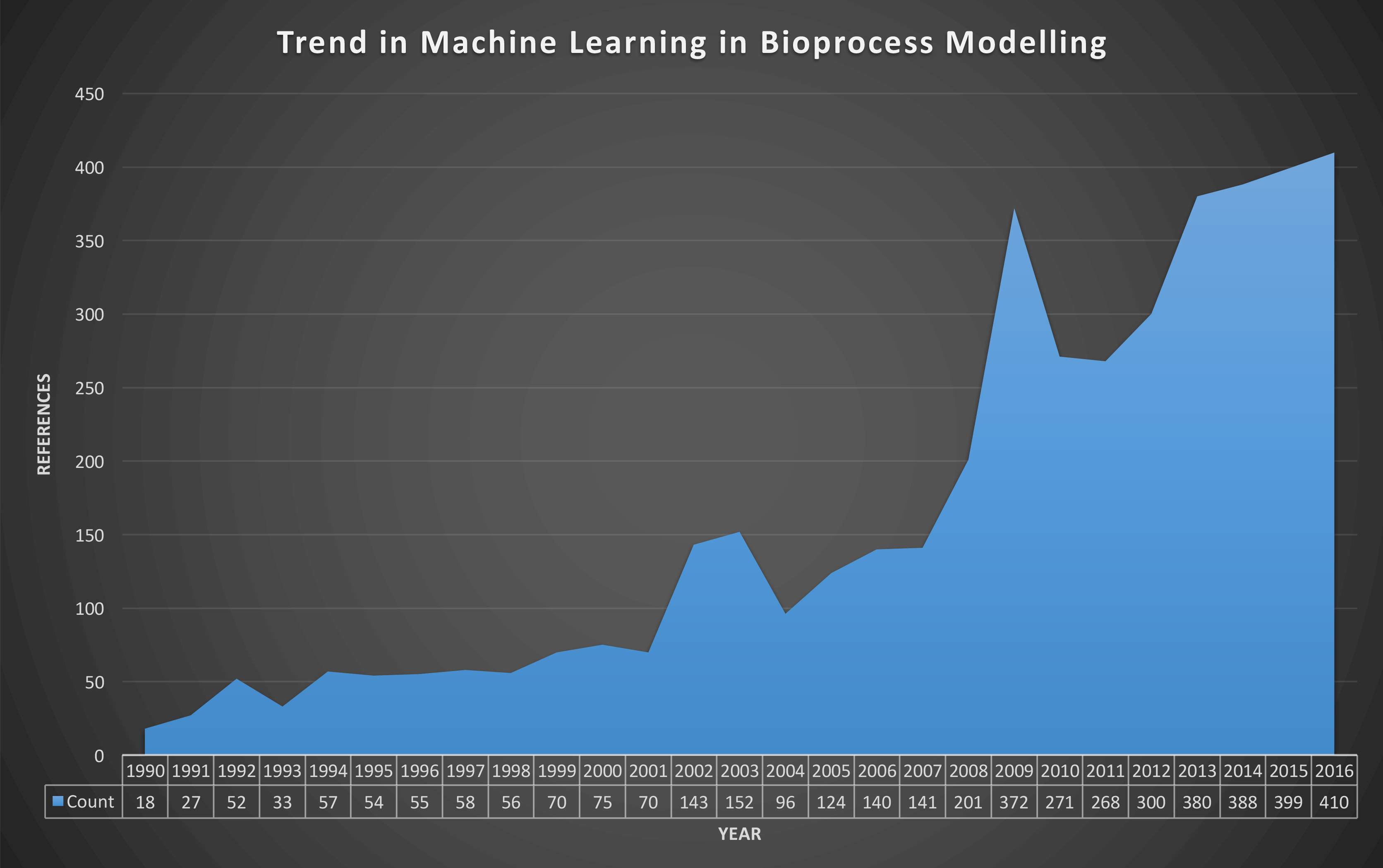

Figure 1: Evolution of the number of peer-reviewed publications resulted from the query “Bioprocess Machine Learning” from Google Scholar [03/10/2017].
Statistical, data-driven or machine-learning models rely on inferences based on collected data. The use of modelling approaches in bioprocessing has increased steadily over more than 20 years (Figure 1). Machine learning models can be subdivided into supervised and unsupervised learning algorithms, depending on the presence or absence of process output data in observations, respectively. Supervised learning algorithms are commonly used for the quantification of CPPs or CQAs and assessing their interdependency, while unsupervised learning algorithms are commonly used in classification applications. Several applications have been reported using these modelling approaches, summarised below:10,12
- Soft sensors – real-time estimation of CPPs / CQAs, based on other real-time measurements and online parameter estimations
- Mode of operation selection – based on the classification of current process measurements into a discrete mode of operation to select appropriated monitor and control mechanisms
- Chemometric models – extract information present on multidimensional spectra into metabolite concentration estimations
- Multivariate data analysis – for the post-mortem analysis of the variation of Process Analytical Technology (PAT) instrumentation measurements in-process (such as identification of golden batch operation conditions)
- Multivariate statistical process control – for process / product state monitoring and failure detection and identification to detect when the process is deviating from its normal / optimal behaviour and source causes
- Process state-observers – estimate the entire process state, based on real-time input variable measurements
- Dynamic simulations – for exploring process parameter settings and optimisation of dynamic operating conditions as well as uncertainty and sensibility analysis
- Closed-loop, adaptive, model-predictive control – predicts the process state for different control actions and acts accordingly.
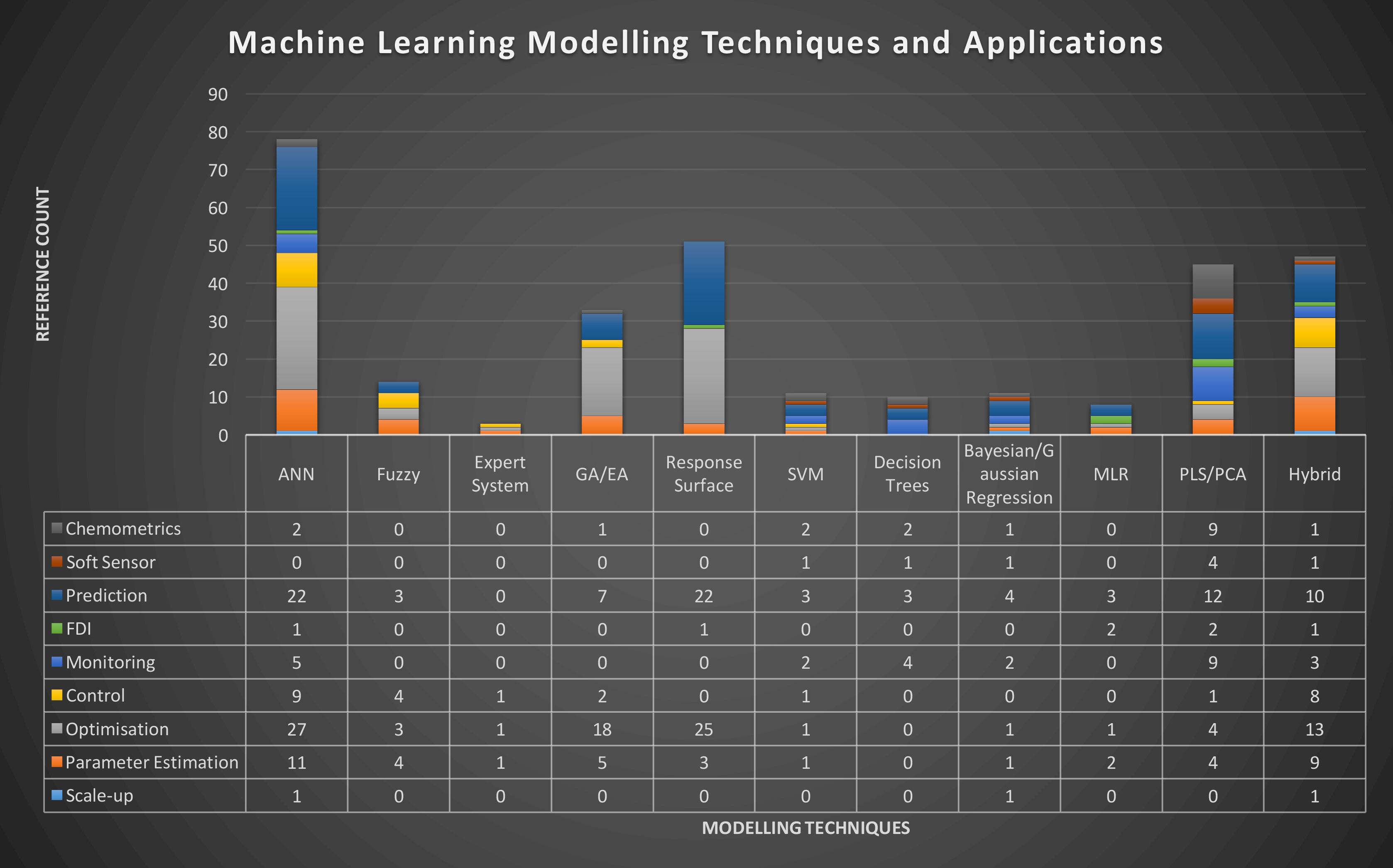

Figure 2: Distribution of different modelling techniques over the most common applications Google Scholar references in bioprocess operation/bioproduct manufacturing for the period [01/01/2017; 03/10/2017]. Modelling techniques and applications were assessed individually for both “bioprocess”, “biopharmaceutics” and “monoclonal antibody” search terms.
Given the current availability of faster and more practical modelling packages, several modelling approaches have been recently applied to biopharmaceutical process / product modelling in the above mentioned areas (Figure 2). Artificial Neural Network (ANN) appears to be the most commonly-used modelling approach, due to its ability to capture non-linear relationships in dynamic systems, as well as to estimate parameters of other models.
Latent Variable methods, such as Partial Least Squares (PLS), may provide better solutions for multivariate regressions of PAT instrumentation data, given their ability to determine correlations between thousands of multi-dimensional variables at once. Recursive partitioning algorithms (or ‘tree’ models) are commonly used as classification models for root-cause analysis.18 However, the majority (>70% – data not shown) of applications found in literature report pattern recognition (classification) applications for early-stage biopharmaceutical product development.
Several disadvantages of data-driven models in biopharmaceutical industry have also been reported.10,18,19 The lack of representative datasets (different batches describing all possible sources of variability) for model development, testing and validating may limit their ability to generalise over wider operability regions. Black-box model parameters are difficult to interpret and, due to the complexity of models, not readily stored and integrated into interpretable knowledge (with rare exceptions).
Furthermore, these models have several disadvantages regarding the uncertainty of predictions outside the calibration space (extrapolation), inside the calibration space when insufficient observations are available for model development (overfitting), and collinearity (correlation-causation) effects, which only become apparent with the introduction of more data to model.10 Another challenge in biopharmaceutical manufacturing process lies in the collection of valid modelling data. Biopharmaceutical manufacturing data include multi-modal, multivariate data, noisy and missing values. The effective pre-processing of raw data into ‘ready-to-model’ data is time-consuming and may introduce artifacts or loss of information. Recently, modelling in the QbD approach was extensively reviewed resulting in several considerations including Good Modelling Practices.4 The main challenges and opportunities for process modelling in biomanufacturing development and operation include:
- Higher transparency of model methodologies and assumptions
- Improved criteria for quantitatively comparing model candidates
- Sensitivity analysis of model predictions with respect to changes in input variables
- Generalised uncertainty analysis strategies, for the quantification prediction error and noise variability effects on model outputs
- Reproducibility, replicability of the results and the stability of the model to data updates
- Feasibility and flexibility analysis, which also takes into consideration the inherent uncertainty of model parameters during the normal process operation to define better control spaces
- Clarity on the explicit integration of domain expertise with black-box modelling approaches (hybrid semi-parametric models as a viable option)
- Dynamic and adaptive DoE for multi-process unit operations
- Scale-independent models and the evaluation of scalability of model parameters across different scales
- Real-time predictions and analysis, to support RTR (Real Time Release) of biotherapeutic drugs.
Although the biopharmaceutical industry may be reluctant to embrace machine learning as a standard tool for bioprocess development – due to the potential catastrophic consequences of faulty products – biopharmaceutical manufacturing is presenting us every year with innovative applications and case studies. The technological progress in machine learning and computing will inevitably lead to broader applicability of these techniques and such case studies are important in providing the community with useful benchmarking material.
References
- Zhang YHP, Sun J, Ma Y. Biomanufacturing: history and perspective. J Ind Microbiol Biotechnol. 2017;44(4–5):773–84.
- FDA. Guidance for Industry PAT: A Framework for Innovative Pharmaceutical Development, Manufacuring, and Quality Assurance. FDA Off Doc. 2004;(September):16.
- U.S. Department of Health and Human Services Food and Drug Administration. ICH Q8(R2) Pharmaceutical Development. Work Qual by Des Pharm. 2009;8(November):28.
- Djuris J, Djuric Z. Modeling in the quality by design environment: Regulatory requirements and recommendations for design space and control strategy appointment. Int J Pharm. 2017;533(2):346–56.
- Neubauer P, Cruz N, Glauche F, Junne S, Knepper A, Raven M. Consistent development of bioprocesses from microliter cultures to the industrial scale. Eng Life Sci. 2013 May;13(3):224–38.
- Kumar V, Bhalla A, Rathore AS. Design of experiments applications in bioprocessing: Concepts and approach. Biotechnol Prog. 2014 Jan;30(1):86–99.
- Mandenius C, Titchener-hooker NJ. Measurement, Monitoring, Modelling and Control of Bioprocesses. Mandenius C-F, Titchener-Hooker NJ, editors. Berlin, Heidelberg, Heidelberg: Springer Berlin Heidelberg; 2013. 285 p. (Advances in Biochemical Engineering/Biotechnology; vol. 132).
- Hausmann R, Henkel M, Hecker F, Hitzmann B. Present Status of Automation for Industrial Bioprocesses. Current Developments in Biotechnology and Bioengineering: Bioprocesses, Bioreactors and Controls. Elsevier Inc.; 2016. 725-757 p.
- You C, Percival Zhang YH. Biomanufacturing by in vitro biosystems containing complex enzyme mixtures. Process Biochem. 2017;52:106–14.
- Solle D, Hitzmann B, Herwig C, Pereira Remelhe M, Ulonska S, Wuerth L, et al. Between the Poles of Data-Driven and Mechanistic Modeling for Process Operation. Chemie-Ingenieur-Technik. 2017 May;89(5):542–61.
- Guo W, Sheng J, Feng X. Mini-review: In vitro Metabolic Engineering for Biomanufacturing of High-value Products. Comput Struct Biotechnol J. 2017;15:161–7.
- Rathore AS, Garcia-Aponte OF, Golabgir A, Vallejo-Diaz BM, Herwig C. Role of Knowledge Management in Development and Lifecycle Management of Biopharmaceuticals. Pharm Res. 2017;34(2):243–56.
- Farzan P, Mistry B, Ierapetritou MG. Review of the important challenges and opportunities related to modeling of mammalian cell bioreactors. AIChE J. 2017 Feb 1;63(2):398–408.
- Mears L, Stocks SM, Sin G, Gernaey KV. A review of control strategies for manipulating the feed rate in fed-batch fermentation processes. J Biotechnol. 2017;245:34–46.
- Sommeregger W, Sissolak B, Kandra K, von Stosch M, Mayer M, Striedner G. Quality by control: Towards model predictive control of mammalian cell culture bioprocesses. Biotechnol J. 2017;12(7):1–7.
- Mears L, Stocks SM, Albaek MO, Sin G, Gernaey KV. Mechanistic Fermentation Models for Process Design, Monitoring, and Control. Trends Biotechnol. 2017 Oct;35(10):914–24.
- St Amand MM, Tran K, Radhakrishnan D, Robinson AS, Ogunnaike BA. Controllability analysis of protein glycosylation in Cho cells. Andersen MR, editor. PLoS One. 2014 Feb 3;9(2):e87973.
- Hill T, Waner A. Utilizing Machine-Learning Capabilities: When Will Big Data Analytics Change Biopharmaceutical and Pharma Manufacturing? Genet Eng Biotechnol News. 2017 Jan 15;37(2):28–9.
- Bate A, Hanif S. A perspective on machine learning in the pharmaceutical industry. 2017.
Biography




Professor Jarka Glassey is currently the Executive Vice President of the European Society of Biochemical Engineering Sciences (ESBES) and Vice President Technical of the IChemE. She is based at Newcastle University and her expertise is in Quality by Design, process analytical technologies and bioprocess modelling. The main emphasis of her research is on using advanced modelling techniques for (bio)process development for biologics and small molecules.
The rest of this content is restricted - login or subscribe free to access
Thank you for visiting our website. To access this content in full you'll need to login. It's completely free to subscribe, and in less than a minute you can continue reading. If you've already subscribed, great - just login.
Why subscribe? Join our growing community of thousands of industry professionals and gain access to:
- bi-monthly issues in print and/or digital format
- case studies, whitepapers, webinars and industry-leading content
- breaking news and features
- our extensive online archive of thousands of articles and years of past issues
- ...And it's all free!
Click here to Subscribe today Login here
Issue
Related topics
Related organisations
European Society of Biochemical Engineering Sciences, Newcastle University