MicroRNAs and their relatives – new avenues in biomedical research
Posted: 23 November 2007 | Dr. Jan Weiler, Novartis Institutes for BioMedical Research, siRNA Therapeutics, Basel, Switzerland | No comments yet
Non-coding RNAs (ncRNAs) consist of a growing heterogeneous class of transcripts defined as RNA molecules that lack any extensive “Open Reading Frame” (ORF) and function as structural, catalytic or regulatory entities rather than serving as templates for protein synthesis. While non-coding sequences make up only a small fraction of the DNA of prokaryotes, among eukaryotes, […]
Non-coding RNAs (ncRNAs) consist of a growing heterogeneous class of transcripts defined as RNA molecules that lack any extensive “Open Reading Frame” (ORF) and function as structural, catalytic or regulatory entities rather than serving as templates for protein synthesis. While non-coding sequences make up only a small fraction of the DNA of prokaryotes, among eukaryotes, the proportion of DNA that does not code for protein increases with their complexity, underscoring the likelihood of a “hidden layer” of gene regulation in animal genomes1.
Although previously being classified as “junk”, recent studies strongly indicate that ncRNAs play central roles in regulating gene expression at multiple levels including; transcription, alternative splicing, RNA modification, such as RNA editing, RNA stability and translation and chromatin modification. Still, there is an ongoing debate on whether ncRNAs are important functional contributors to an organism’s complexity or rather represent transcriptional “background” or “noise”2. NcRNAs can be subdivided into two main classes: “housekeeping” RNAs such as transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), small nuclear RNAs (snRNAs) and regulatory RNAs (“riboregulators”), such as small nucleolar RNAs (snoRNAs), and microRNAs (miRNAs), which are expressed at certain stages of organism development or cell differentiation and can modulate expression of other genes on the levels of transcription or translation. In particular, miRNAs represent a class of ncRNAs which has recently attracted major attention within the scientific community. Although information on the function of miRNAs is still sparse, an increasing body of evidence suggests that these molecules play major roles in important biological processes and may contribute to the pathogenesis of human diseases3. Consequently, an increasing number of research scientists who have traditionally worked on proteins, are beginning to move their focus towards studying the world of these tiny RNA molecules. The potential and prospects of miRNAs and other newly discovered ncRNA family members as a new class of therapeutic are reviewed, and the constraints associated with current miRNA target validation approaches are discussed.
MiRNA biology and pathology
MiRNAs are short (17-25 nt), endogenously expressed RNAs which either reside in introns or exons of coding genes or are found in “intergenic genomic regions”, which are defined as DNA sequences being located > 1 kb away from any annotated or predicted genes4. Some miRNA genes are solitary, and are expressed under the control of their own promoters, while others are arranged in clusters and may be transcribed as paired or polycistronic transcripts. MiRNA biogenesis is a multi-step process, which has been extensively investigated5. MiRNAs are generated from hairpin-like RNAs known as precursor miRNAs (pre-miRNAs) which are transported from the nucleus to the cytoplasm where they are digested by the dsRNA-specific ribonuclease III enzyme Dicer to release the mature miRNA. The excised miRNA is incorporated into the so-called “miRNA-containing ribonucleoprotein particle” (miRNP), a complex similar to the RNA-induced silencing complex (RISC), which participates in the RNA interference process. The complex bound miRNA binds specific mRNA targets and, depending on the degree of complementarity, interferes with the translational machinery or induces degradation of the associated mRNA targets by a mechanism not fully understood6.
Studies in multiple model organisms have shown that an animal without miRNAs cannot live or reproduce and it is well appreciated that these molecules are critically involved in a variety of important biological processes, such as developmental timing and early patterning, cell proliferation and differentiation, apoptosis, fat metabolism, tissue morphogenesis (reviewed in reference 7) and response to environmental stress8. Given the importance of miRNA-mediated regulation, it is obvious that perturbation of the miRNA silencing machinery could have major functional consequences including an increased susceptibility to disease phenotypes. Indeed, as highlighted by a growing number of publications, miRNAs are increasingly believed to be involved in the pathology of human diseases including cancer, neurodegenerative diseases, metabolic disease, and viral infection (Table 1).
MiRNAs could contribute to disease phenotype through a variety of mechanisms7. MiRNA expression levels can be directly altered due to genomic rearrangements affecting the identity or expression of miRNAs or core components of the miRNA machinery. Alternatively, a gain or loss of a miRNA-target interaction may occur due to mutations in the miRNA gene itself or in the 3’-UTR of a miRNA target gene. While there are numerous reports on the contribution of miRNA dysregulation to disease, in particular cancer20, an intriguing example for a phenotype associated with a perturbation of a miRNA-target interaction, is the identification of the molecular basis of meatiness in the Texel sheep21. Combination of a quantitative trait loci (QTL) scan with a close genetic analysis revealed a point mutation in the 3’-UTR of the muscle-specific gene myostatin. This mutation creates an illegitimate miRNA binding site for mir-1 and mir-206, two miRNAs highly enriched in skeletal muscle. Additional experiments suggest that this new miRNA binding site induces mir-1 and mir-206-dependent downregulation of myostatin, resulting in the build up of extra muscle fibres. A recent study on the identification of naturally occurring small nuclear polymorphisms (SNPs) suggests many polymorphisms in miRNA target sites although phenotypic effects of these SNPs remain to be determined22.
Tools in miRNA target discovery
MiRNA and other ncRNA genes remained undetected for many years, mainly due to the difficulty to computationally identify them and the lack of sensitivity in the techniques to experimentally detect them. The discovery of siRNAs and miRNAs as new players participating in the cell’s control system has triggered a major race and excitement in the development of computational approaches and experimental platforms aimed at the systematic identification and functionalisation of new ncRNA species. Integrative genomics efforts, combining computational predictions with large-scale cloning and microarray expression analysis, have recently enabled a major expansion of the list of miRNA candidates in animals23,24.
MiRNA detection and expression profiling
The high degree of conservation and the fact that many miRNAs are differentially expressed indicates the relevance of miRNAs in critical biological processes but also suggests that they might be useful as diagnostic or prognostic markers. According to a recent study, miRNA expression signatures can be more accurate in tumor classification than classification based on mRNA profiles25. Clearly, the ability to experimentally determine the spatial and temporal expression of miRNAs is a critical means to establish their role in gene regulation under normal or diseased conditions. Over recent years, researchers in academic groups as well as in biotech companies have invested heavily in efforts to develop technologies to experimentally assay miRNAs and other ncRNAs in biological samples (“RNomics”). Several miRNA expression profiling platforms have been developed, the most commonly applied technologies being microarrays26-36, RT-PCR37,38 or large-scale cloning and sequencing approaches39. A major advantage of microarray-based approaches is the possibility to assay large numbers of molecules simultaneously with high-throughput. Thus, as the number of discovered ncRNAs is growing, microarrays are likely to become the method of choice for global expression profiling studies. Despite the advancement in this field, there is considerable disagreement with respect to miRNA expression patterns for a specific cell-type, which at least in part may depend on the individual technical differences between the platforms40. Thus, efforts to standardise miRNA profiling protocols and to establish thoroughly characterised reference sets are of paramount importance. In addition, because of the high costs and efforts involved in follow-up experiments on individual miRNA candidates, validation of putative expression differences by complementary methods such as PCR-based technologies which offer greater sensitivity and specificity remains advisable28. Finally, in situ hybridisation technologies41 are critical to complement primary expression data and to establish the specificity and cellular resolution of miRNA expression patterns in vivo.
MiRNA target validation – computational approaches
The fact that a miRNA has been found to be abnormally expressed, for example, in cancer tissues, does not prove that these molecules play a causal role in disease pathogenesis as opposed to a rather consequential or compensatory role. Nevertheless, dysregulation of miRNAs during certain disease states may serve as a starting point for a more in depth study aiming at further validation of these entities as potential biomarkers or pharmaceutical targets. In subsequent studies, the mechanistic role of the miRNA target candidates needs to be established. However, to elucidate the mRNA targets modulated by a specific miRNA is a fundamental problem in miRNA research. In animals, miRNAs bind to the 3’-UTR regions of their targets with imperfect complementarity but the precise mechanism of miRNA-directed protein inhibition has remained largely unclear6. Current computational algorithms to predict putative miRNA-target recognition sites rely on base pairing between the “seed”, comprising the 5’-end of a miRNA spanning bases two-eight and its target mRNAs. However, recent studies strongly suggest that the physiological functionality of a certain miRNA-binding site is likely to depend on additional factors other than pure “seed” sequence complementary, such as binding sites for other cofactors such as miRNA binding proteins42,43 as well as the distance between individual miRNA binding sites44 and structural accessibility of the target site45,46. In addition, a recent publication47 suggested that mRNAs can also be negatively regulated by miRNA binding sites located in the 5’-UTR or even within the coding region. Finally, most of the currently available miRNA target prediction algorithms do not take into account that a miRNA-target interaction requires spatial and temporal co-expression of miRNA and its putative targets. Thus, the number of targets controlled by miRNAs on single cell level will be considerably smaller than one would predict by transcriptome-wide searching for mere individual seed matches48. As a consequence, most of the commonly used miRNA target prediction algorithms tend to either overestimate the number of miRNA targets or, if evolutionary conservation is applied, yield a significant number of false negatives49. On the other hand, the current lack of experimental data demonstrating the interaction of specific miRNAs with their respective mRNA targets in vivo represents a serious limitation to develop accurate algorithms for miRNA target prediction.
MiRNA target validation – experimental approaches
Confirming miRNA predictions should be ideally performed experimentally by matching miRNA changes with protein abundance or activity50. Since such proteomic approaches are challenging for technical reasons, some researchers have attempted to correlate mRNA microarray with miRNA expression data in order to analyse the effects of miRNAs on target mRNA expression51-53. One limitation of this approach is that it will only pick up target candidates where a significant degree of mRNA degradation is induced upon miRNA interaction, while cases where translational inhibition takes place without triggering a significant degree of RNA degradation may not be visible in classical microarray approaches. Finally, mRNA expression profiles obtained upon artificial manipulation of miRNA levels might cause artifacts and do not allow one to distinguish between direct miRNA-target effects and indirect effects such as downstream consequences of target manipulation. Biochemical approaches, such as miRNP immunopurification, which identify functional miRNA target candidates based on their physical interaction with miRNAs in vitro, may be useful to complement microarray-based approaches54.
Although, miRNAs are estimated to regulate the expression of a large number of protein-encoding genes (at least 30%) in the human genome, there are only a few cases reported where the physiological function of a mammalian miRNA has been elucidated in vivo. Today, only few mammalian miRNA knockouts have been reported. In Dicer-deficient mice, processing of precursor miRNAs to the mature miRNAs is stalled, leading to impaired embryonic angiogenesis and embryonic death. On the other hand, conditional Dicer inactivation indicates that miRNAs are essential in skin, lung epithelium and vertebrate limp morphogenesis7. Mice lacking mir-1-2, a miRNA strongly enriched in cardiac tissue and skeletal muscle displayed severe cardiac abnormalities55. Mice deficient in the heart-enriched mir-208 failed to induce an apparent phenotype within the first month of age but failed to undergo cardiac remodeling when they were subjected to cardiac stress14, suggesting that miRNA-mediated regulation may play an important role during cardiac development and physiology. Finally, recent studies on mir-155 knockout mice suggest that miRNAs may play a role in regulating homeostasis and function in the immune system56,57.
MiRNAs in drug discovery
The most obvious strategy to interfere with disease-relevant miRNAs is to use antisense oligonucleotides complementary to the corresponding miRNA sequence or its precursor such that the miRNA will be blocked from binding to its targets (see Figure 1). Recently, two studies conducted by independent groups using chemically modified anti-miRNA oligonucleotides (AMOs)58 have demonstrated the concept of effective miRNA downregulation in vivo. Administration of 2’-O-Methoxyethyl phoshorothioate59, respectively cholesterol conjugated, 2’-O-Methyl modified oligonucleotides (“antagomirs”)60 specifically targeting the liver specific miRNA mir-122 resulted in a decrease in plasma cholesterol levels and a reduced rate of cholesterol biosynthesis in mice models. On the other hand, restoring miRNA function by the exogenous delivery of a synthetic miRNA mimic such as an siRNA-like dsRNA may have therapeutic benefit, in particular in certain cancers where tumor-suppressor miRNAs are underrepresented or missing.
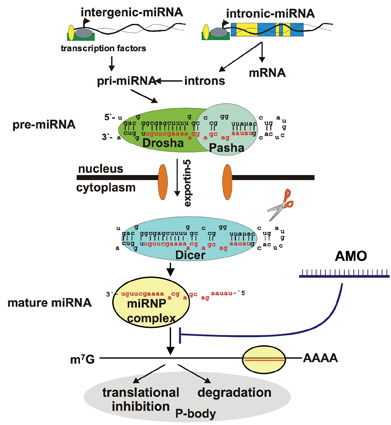

Figure 1: Principle of blocking a mature miRNA from binding to its mRNA targets using synthetic anti-mir oligonucleotides (AMOs)
Certainly, the progress in the area of miRNA-based medicines is closely associated with the ongoing efforts aiming at the development of RNA interference (RNAi) therapies, designed to specifically turn off the activity of disease-associated genes. Developing both RNAi and miRNA therapeutics face several critical obstacles which comprise silencing efficacy and specificity, stability of the inhibitors in the body, and most importantly delivery to and uptake by the organ or cells of interest. Although, first siRNA studies performed in animals look encouraging61, the above limitations have to be overcome before these approaches can be translated into clinically viable options for humans.
Alternative approaches to therapeutically modulate miRNAs without using Watson-Crick base-pairing such as screening for or rationally designing low-molecular-weight RNA-ligands will be challenging due to specificity issues62.
The question remains whether small RNAs such as miRNAs represent suitable target structures for therapeutic intervention in humans. Many of these tiny regulators are believed to act in a highly tissue or even cell type specific manner. Furthermore, a significant number of miRNAs appear to be co-expressed as transcription units and may act in a combinatorial manner. Thus, targeting a subset of different miRNAs might be necessary to significantly modulate one given mRNA target. On the other hand, in cases where one single miRNA regulates multiple functionally related genes, one miRNA inhibitor modulating a whole disease-relevant pathway by reducing several proteins at once may be more effective than the more traditional agents tailored to target one single protein63. In each individual case, efficacy, miRNA pathway redundancy, specificity and potential adverse side-effects remain to be carefully assessed in vivo. In this regard, miRNAs critical in viral diseases may represent the most promising target class at this point in time64.
Future prospects
Although, after several years of intense research in the field, we know a lot about how miRNAs are made, there is still much left to learn about the mechanistic aspects of the ncRNA machinery in general. Currently, there are more than 500 mammalian miRNAs listed in the latest release (version 10.0) of the miRBase65 and it is expected that this number will exceed the thousand mark23. While most of the known miRNAs are evolutionarily conserved, several lines of evidence suggest that there remain large numbers of miRNAs yet to be identified which may be human specific. At the same time, other ncRNA species such as piRNA (piwi-interacting RNAs), rasiRNAs (repeat-associated small interfering RNAs and other endogenous siRNAs3 are emerging, the roles of which are far from being fully characterised. An increasing number of publications suggest an important role for miRNAs in specific physiological pathways underlying the disease pathology of multiple human diseases.
So, what needs to be done to fill up the miRNA drug discovery tool box? First, computational approaches in combination with systematic experimental verification remain essential to complete the repository of miRNAs and to identify the transcription factors that control their expression. In addition, technologies to quantify small RNAs will become a pivotal tool in miRNA-related research. There is a growing consensus in the scientific community of the need for a more comprehensive repository cataloging miRNA expression data38,66 generated from specific tissues and cell types in physiological and pathologic conditions, analogous to existing databases for storing information on mRNA expression profiles. In parallel, efforts to systematically identify and catalogue polymorphisms mapping to human miRNAs, as well as at miRNA target sites67 may contribute to the identification of further disease associated miRNA candidates. Furthermore, the generation of a larger set of experimentally validated miRNAs targets is critical in order to improve and refine the currently available miRNA target prediction algorithms. Finally, characterised tools to induce miRNA gain- and loss-of function in appropriate animal models remain essential for identifying the significance of miRNAs for a specific disease process.
As systematic genomic and proteomic analysis have contributed to defining the molecular network of diseases, it is increasingly appreciated that small RNA entities such as miRNA but also other ncRNA products may be involved in structural, functional and regulatory interactions at multiple levels and thus yield additional information on the biology of human diseases. In fact, these molecules previously degraded to genomic “junk” may be as important to evolution68 and human health and illness as genes themselves. In light of such considerations, ncRNA research will almost certainly become an integral part of the Systems Biology initiative and could provide a much deeper understanding of regulatory pathways in normal as well as diseased tissue. Such studies will hopefully lead to the identification of innovative avenues for therapeutic intervention. Proof-of-principle studies for the therapeutic application of RNAi-based61 as well as antisense therapeutics69 are underway in different disease indications. Success of even one of these approaches will certainly provide fruitful ground and motivation for further investigation of the potential of miRNAs, and their relatives, to combat human disease.
Acknowledgements
This article is dedicated to my thesis advisor Professor Wolfgang Pfleiderer on the occasion of his 80th birthday. I would like to thank Dr. William Wishart for critically reading the manuscript.
References
- Mattick, J.,S. The functional genomics of noncoding RNA. Science 2005, 309: 1527-1528.
- Ponjavic, J., Ponting, C., P., Lunter, G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007, 17: 556-565.
- Ambros, V., Chen, X., The regulation of genes and genomes by small RNAs. Development 2007, 134: 1635-1641.
- Nary, K.,V. MicroRNA biogenesis: Coordinated cropping and dicing. Nature Reviews Molecular Cell Biology 2005, 65: 376-385.
- Du, T., Zamore, P.,D. MicroPrimer: The biogenesis and function of microRNA. Development 2005, 132: 4645-4652.
- Pillai, R.S., Bhattacharyya, S,N., Filipowicz, W. Repression of protein synthesis by miRNAs: How many mechanisms? Trends in Cell Biology 2007, 173: 118-126.
- Kloosterman, W.P., Plasterk, R.A. The diverse functions of microRNAs in animal development and disease. Developmental Cell 2006, 11: 441-450.
- Leung, A.K.L., Sharp, P.A. microRNAs: a safeguard against turmoil? Cell 2007, 130: 581-585.
- Calin, G.A, Ferracin, M., Cimmino, A., Di Leva, G., Shimizu, M., Wojcik, S.E., Iorio, M.V., Visone, R., Sever, N.I., Fabbri, M., Iuliano, R., Palumbo, T., Pichiorri, F., Roldo, C., Garzon, R., Sevignani, C., Rassenti, L., Alder, H., Volinia, S., Liu, C.G, Kipps, T.J., Negrini, M., Croce, C.M. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. New England J. Med. 2005, 35317: 1793-1801.
- He, L., Thomson, J.M, Hemann, M.T., Hernando-Monge, E., Mu, D., Goodson, S., Powers, S., Cordon-Cardo, C., Lowe, S.W., Hannon, G.J., Hammond, S.M. A microRNA polycistron as a potential human oncogene. Nature 2005, 435: 828-833.
- Shell, S., Park, S.M, Radjabi, A.R, Schickel, R. Kistner, E.O., Jewell, D.A., Feig, C., Lengyel, E., Peter, M.E.. Let-7 expression defines two differentiation stages of cancer. Proc. of the Natl. Acad. Sci. USA 2007, 104: 11400-11405.
- Ma, L., Teruya-Feldstein, J., Weinberg, R.A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 2007, 449: 682-688.
- Poy, M., Eliasson, L., Krutzfeldt, J., Kuwajima, S., Ma, X., MacDonald, P.E., Pfeffer, S., Tuschl, T., Rajewsky, N., Rorsman, P., Stoffel, M. A pancreatic islet-specific microRNA regulates insulin secretion. Nature 2004, 432: 226-230.
- Care A., Catalucci D., Felicetti F., Bonci D., Addario A., Gallo P., Bang M.L., Segnalini P., Gu Y., Dalton N.D, Elia L., Latronico M.V.G, Hoydal M., Autore C., Russo M.A, Dorn G.W, 2nd, Ellingsen O., Ruiz-Lozano P., Peterson K.L, Croce C.M, Peschle C., Condorelli G. MicroRNA-133 controls cardiac hypertrophy. Nature medicine 2007, 13: 613-618.
- van Rooij, E., Sutherland, L.B., Qi, X., Richardson, J.A., Hill, J., Olson, E.N. Control of Stress-Dependent Cardiac Growth and Gene Expression by a MicroRNA. Science 2007, 316: 575-579.
- Sonkoly, E., Wei, T., Janson, P.C.J., Saeaef, A., Lundeberg, L, Tengvall-Linder, M., Norstedt, G., Alenius, H., Homey, B., Scheynius, A., Stahle, M., Pivarcsi, A.. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS One 2007, 2:1-8.
- Abelson, J.F., Kwan, K.Y., O’Roak, B.J., Baek, D.Y., Stillman, A.A., Morgan, T.M., Mathews, C.A., Pauls, D.L., Rasin, M., Gunel, M., Davis, N.R., Ercan-Sencicek, A., Gulhan, G., Danielle H., Spertus, J.A., Leckman, J.F., Dure, L.S., IV, Kurlan, R., Singer, H.S., Gilbert, D.L., Farhi, A., Louvi, A., Lifton, R.P., Sestan, Ne., State, M.W. Sequence Variants in SLITRK1 Are Associated with Tourette’s Syndrome. Science 2005, 310: 317-320.
- Kim, J., Inoue, K., Ishii, J., Vanti, W.B., Voronov, S.V., Murchison, E., Hannon, G., Abeliovich, A. A MicroRNA Feedback Circuit in Midbrain Dopamine Neurons. Science 2007, 317: 1220-1224.
- Jopling, C.L., Yi, M., Lancaster, A.M., Lemon, S.M., Sarnow, P. Modulation of Hepatitis C Virus RNA Abundance by a Liver-Specific MicroRNA. Science 2005, 309: 1577-1581.
- Calin, G.A., Croce, C.M. Chromosomal rearrangements and microRNAs: a new cancer link with clinical implications. J. Clin. Invest. 2007, 117(8), 2059-2066.
- Clop, A., Marcq, F., Takeda, H., Pirottin, D., Tordoir, X, Bibe, B., Bouix, J., Caiment, F., Elsen, J.M, Eychenne, F., Larzul, C., Laville, E., Meish, F., Milenkovic, D., Tobin, J., Charlier, C., Georges, M.. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nature Genet. 2006, 38: 813-818.
- Saunders, M.A., Liang, H., Li, W.H.. Human polymorphism at microRNAs and microRNA target sites. Proc. Natl. Sci. USA 2007, 104: 3300-3305.
- Bentwich, I., Avniel, A., Karov, Y., Aharonov, R., Gilad, S., Barad, O., Barzilai, A., Einat, P., Einav, U., Meiri, E., Sharon, E., Spector, Y., Bentwich, Z. Identification of hundreds of conserved and nonconserved human microRNAs. Nature Genet. 2005, 37, 766-770.
- Berezikov, E., van Tetering, G., Verheul, M., van de Belt, J., van Laake, L., Vos, J., Verloop, R., van de Wetering, M., Guryev, V., Takada, S., van Zonneveld, A.J., Mano, H., Plasterk, R., Cuppen, E. Many novel mammalian microRNA candidates identified by extensive cloning and RAKE analysis. Genome Res. 2006, 16: 1289-1298.
- Lu, J., Getz, G., Miska, E.A., Alvarez-Saavedra, E., Lamb, J., Peck, D., Sweet-Cordero, A., Ebert, B.L., Mak, R.H., Ferrando, A.A., Downing, J.R., Jacks, T., Horvitz, H., Golub, T.R. MicroRNA expression profiles classify human cancers. Nature 2005, 435: 834-838.
- Castoldi, M., Benes, V., Hentze, M.W., Muckenthaler, M.U. miChip: A microarray platform for expression profiling of microRNAs based on locked nucleic acid (LNA) oligonucleotide capture probes. Methods 2007, 43: 146-152.
- Wang, H., Ach, R.A., Curry, B. Direct and sensitive miRNA profiling from low-input total RNA. RNA 2007, 13: 151-159.
- Beuvink, I., Kolb, F.A., Budach, W., Garnier, A., Lange, J., Natt, F., Dengler, U., Hall, J., Filipowicz, W., Weiler, J.. A novel microarray approach reveals new tissue-specific signatures of known and predicted mammalian microRNAs. Nucl. Acids Res. 2007, 35: e52/1-e52/11.
- Shingara, J., Keiger, K., Shelton, J., Laosinchai-Wolf, W., Powers, P., Conrad, R., Brown, D., Labourier, E. An optimized isolation and labeling platform for accurate microRNA expression profiling, RNA 2005, 11: 1461–1470 .
- Monticelli, S, Ansel, K.M, Xiao, C, Socci, N.D, Krichevsky, A.M, Thai, T.H, Rajewsky, N, Marks, D.S, Sander, C, Rajewsky, K., Rao, A., Kosik, K.S. MicroRNA profiling of the murine hematopoietic system, Genome Biol. 2005, 6: R71
- Nelson, P.T, Baldwin, D.A, Scearce, L.M, Oberholtzer, J.C, Tobias, J.W, Mourelatos, Z. Microarray-based, high-throughput gene expression profiling of microRNAs, Nat. Methods 2004, 1: 155–161.
- Liang, R.Q, Li, W, Li, Y, Tan, C.Y, Li, J.X, Jin, Y.X, Ruan, K.C. An oligonucleotide microarray for microRNA expression analysis based on labeling RNA with quantum dot and nanogold probe 2005, Nucleic Acids Res., 33:e17.
- Miska, E.A, Alvarez-Saavedra, E., Townsend, M., Yoshii, A., Sestan, N., Rakic, P., Constantine-Paton, M., Horvitz, H.R. Microarray analysis of microRNA expression in the developing mammalian brain Genome Biol. 2004, 5: R68.
- Sun, Y., Koo, S., White, N., Peralta, E., Esau, C., Dean, N.M, Perera, R.J. Development of a micro-array to detect human and mouse microRNAs and characterization of expression in human organs Nucleic Acids Res. 2004, 32, e188.
- Barad, O., Meiri, E., Avniel, A., Aharonov, R., Barzilai, A., Bentwich, I., Einav, U., Gilad, S., Hurban, P., Karov, Y., Lobenhofer, E.K., Sharon, E., Shiboleth, Y.M., Shtutman, M., Bentwich, Z., Einat, P.MicroRNA expression detected by oligonucleotide microarrays: system establishment and expression profiling in human tissues, Genome Res. 2004, 14: 2486–2494 .
- Liu, C.G., Calin, G.A, Meloon, B., Gamliel, N., Sevignani, C., Ferracin, M., Dumitru, C.D., Shimizu, M., Zupo, S., Dono, M., Alder H., Bullrich, F., Negrini, M., Croce, C.M. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues Proc. Natl. Acad. Sci. USA 2004, 101: 9740–9744.
- Schmittgen, T.D., Jiang, J., Liu, Q., Yang, L., A high-throughput method to monitor the expression of microRNA precursors, Nucl. Acids Res. 2004, 32: e43/1-e43/10.
- Chen, C, Ridzon, D.A., Broomer, A.J., Zhou, Z., Lee, D.H., Nguyen, J.T., Barbisin, M., Xu, N.Lan, Mahuvakar, V.R., Andersen, M.R., Lao, K.Q, Livak, K.J., Guegler, K.J. Real-time quantification of microRNAs by stem-loop RT-PCR, Nucl. Acids Res., 2005, 33: e179/1-e179/9.
- Landgraf, P., Rusu, M., Sheridan, R., Sewer, A., Iovino, N., Aravin, A., Pfeffer, S., Rice, A., Kamphorst, A.O., Landthaler, M., Lin, C., Socci, N.D., Hermida, L., Fulci, V., Chiaretti, S., Foa, R., Schliwka, J., Fuchs, U., Novosel, A., Muller, R.U, Schermer, B., Bissels, U., Inman, J., Phan, Q., Chien, M., Weir, D.B., Choksi, R., De Vita, G., Frezzetti, D., Trompeter, H.I, Hornung, V., Teng, G., Hartmann, G., Palkovits, M., Di Lauro, R., Wernet, P., Macino, G., Rogler, C.E., Nagle, J.W., Ju, J., Papavasiliou, F.N., Benzing, T., Lichter, P., Tam, W., Brownstein, M.J., Bosio, A., Borkhardt, A., Russo, J.J., Sander, C., Zavolan, M., Tuschl, T., A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007, 129: 1401-1414.
- Jay, C., Nemunaitis, J., Chen, P., Fulgham, P., Tong, A.W. miRNA profiling for diagnosis and prognosis of human cancer. DNA and Cell Biology 2007, 26, 293-300.
- Obernosterer, G., Martinez, J., Alenius, M.. Locked nucleic acid-based in situ detection of microRNAs in mouse tissue sections. Nat. Protocols 2007, 2: 1508-1514.
- Grimson, A., Farh, K.KH., Johnston, W.K., Garrett-Engele, P., Lim, L.P., Bartel, D.P., MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell, 2007, 27, 91-105.
- Didiano, D., Hobert, O. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat. Struct. Mol. Biol., 2006, 13,: 849-851.
- Saetrom, P., Heale, B.S.E., Snove, O.Jr., Aagaard, L., Alluin, J., Rossi, J.J. Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucl. Acids Res., 2007, 35: 2333-2342.
- Long, D., Lee, R., Williams, P., Chan, C.Y, Ambros, V, Ding, Y. Potent effect of target structure on microRNA function. Nat. Struct. Mol. Biol., 2007, 14: 287-294.
- Kertesz, M., Iovino, N., Unnerstall, U., Gaul, U., Segal, E.. The role of site accessibility in microRNA target recognition, Nat. Genetics, 2007, 39: 1278-1284.
- Lytle, J.R., Yario, T.A., Steitz, J.A. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc. Natl. Ac. Sc. 2007, 104: 9667-9672.
- Schmitter, D., Filkowski, J., Sewer, A., Pillai, R.S., Oakeley, E.J., Zavolan, M., Svoboda, P., Filipowicz, W. Effects of Dicer and Argonaute down-regulation on mRNA levels in human HEK293 cells. Nucl. Acids Res., 2006, 34: 4801-4815.
- Maziere, P., Enright, A.J. Prediction of microRNA targets. Drug Discovery Today, 2007, 12: 452-458.
- Vinther, J., Hedegaard, M.M., Gardner, P.P., Andersen, J.S., Arctander, P. Identification of miRNA targets with stable isotope labeling by amino acids in cell culture. Nucl. Acids Res. 2006, 34: e107/1-e107/6.
- Lim, L.P., Lau, N.C., Garrett-Engele, P., Grimson, A., Schelter, J.M., Castle, J., Bartel, D.P., Linsley, P.S., Johnson, J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005, 433: 769-773.
- Wang, X., Wang, X. Systematic identification of microRNA functions by combining target prediction and expression profiling. Nucl. Acids Res. 2006, 34: 1646-1652.
- Sood, P., Krek, A., Zavolan, M., Macino, G., Rajewsky, N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc. Natl. Acad. Sci. USA 2006, 103: 2746-2751.
- Easow, G., Teleman, A.A., Cohen, S.M. Isolation of microRNA targets by miRNP immunopurification. RNA, 2007, 13, 1198-1204.
- Zhao, Y., Ransom, J.F., Li, A., Vedantham, V., von Drehle, M., Muth, A.N., Tsuchihashi, T., McManus, M.T., Schwartz, R.J., Srivastava, D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell, 2007, 129: 303-317.
- Thai, T.H, Calado, D.P., Casola, S., Ansel, K. M., Xiao, C., Xue, Y., Murphy, A. Frendewey, D., Valenzuela, D., Kutok, J.L., Schmidt-Supprian, M., Rajewsky, N., Yancopoulos, G., Rao, A., Rajewsky, K.. Regulation of the Germinal Center Response by MicroRNA-155. Science, 316: 604-608.
- Rodriguez, A., Vigorito, E., Clare, S., Warren, M.V., Couttet, P., Soond, D.R., van Dongen, S., Grocock, R.J., Das, P.P., Miska, E.A., Vetrie, D., Okkenhaug, K., Enright, A.J., Dougan, G., Turner, M., Bradley, A.. Requirement of bic/microRNA-155 for Normal Immune Function. Science, 2007, 316: 608-611.
- Weiler, J., Hunziker, J., Hall, J. Anti-miRNA oligonucleotides (AMOs): ammunition to target miRNAs implicated in human disease? Gene Ther. 2006, 13: 496-502.
- Esau, C., Davis, S., Murray, S.F., Yu, X.X., Pandey, S.K., Pear, M., Watts, L., Booten, S.L., Graham, M., McKay, R., Subramaniam, A., Propp, S., Lollo, B.A., Freier, S., Bennett, C.F., Bhanot, S., Monia, B.P. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab., 2006, 3: 87-98.
- Kruetzfeldt, J., Rajewsky, N., Braich, R., Rajeev, K.G., Tuschl, T., Manoharan, M., Stoffel, M.. Silencing of microRNAs in vivo with ‘antagomirs’. Nature, 2005, 438: 685-689.
- de Fougerolles, A., Vornlocher, H., Maraganore, J., Lieberman, J.. Interfering with disease: a progress report on siRNA-based therapeutics. Nat. Rev. Drug Discovery 2007, 6: 443-453.
- Lagoja, I.M., Herdewijn, P.. Use of RNA in drug design. Expert Opinion on Drug Discovery, 2007, 2: 889-903.
- Mack, G.S. MicroRNA gets down to business. Nat. Biotech. 2007, 25: 631-638.
- Dykxhoorn, D.M.: MicroRNAs in Viral Replication and Pathogenesis. DNA and Cell Biology 2007, 26: 239-249.
- Griffiths-Jones S., Grocock R.J, van Dongen S., Bateman A., Enright A.J, Nucl. Acids. Res. 2006, 34, Database Issue: D140-D144
- Liang, Y., Ridzon, D., Wong, L., Chen, C. Characterization of microRNA expression profiles in normal human tissues. Molecular and Cell Biology- BMC Genomics 2007: 1-20.
- Georges, M., Coppieters, W., Charlier, C. Polymorphic miRNA-mediated gene regulation: contribution to phenotypic variation and disease. Current Opinion in Genetics & Development 2007, 17: 166-176.
- Pollard, K.S., Salama, S.R., Lambert, N., Lambot, M.A., Coppens, S., Pedersen, J.S., Katzman, S., King, B., Onodera, C., Siepel, A., Kern, A.D., Dehay, C., Igel, H., Ares, M.Jr., Vanderhaeghen, P., Haussler, D, An RNA gene expressed during cortical development evolved rapidly in humans. Nature 2006, 443: 167-172.
- Potera, C. Antisense – down, but not out. Nat. Biotech. 2007, 25: 497-499.
Jan Weiler
Novartis Institutes for BioMedical Research, siRNA Therapeutics, Basel, Switzerland
Jan Weiler is a Senior Research Investigator within the siRNA Therapeutics group in the Biologics Center department at the Novartis Institutes for BioMedical Research (NIBR) in Basel, Switzerland. He received his Ph.D. in Organic Chemistry from the University of Konstanz, Germany in the lab of Prof. Dr. Dr. h.c. W. Pfleiderer and was a postdoctoral fellow in the department of Functional Genome Analysis at the German Cancer Research Center DKFZ with Dr. J. Hoheisel. Here his work focused on developing novel microarray technologies. Since he joined Novartis as a labhead in 1997, he has participated in and led antisense-, RNAi- and most recently miRNA related projects to help prioritising novel pharmaceutical targets for drug discovery.




