Turn back the clock and be healed Induced pluripotent stem cells and their future impact on drug discovery and regenerative medicine
Posted: 9 May 2010 |
They are only four years old and are getting everyone very excited; they were Science Magazine’s ‘Breakthrough of the Year 2008’ and Nature’s ‘Method of the Year 2009.’ Their discoverer, Shinya Yamanaka, shared the Lasker Award last year and is no doubt touted for a future Nobel Prize. ‘They’ are induced pluripotent stem cells (or iPS for short). The discovery was that somatic cells from the adult body, whether from a hair, skin biopsy, cord-blood or even adipose tissue, can quite readily be changed back into pluripotent stem cells – ostensibly the state they were in shortly after conception – in the process, erasing the epigenetic modifications that make a brain cell different from, say, a liver cell.

Figure 1
They are only four years old and are getting everyone very excited; they were Science Magazine’s ‘Breakthrough of the Year 2008’ and Nature’s ‘Method of the Year 2009.’ Their discoverer, Shinya Yamanaka, shared the Lasker Award last year and is no doubt touted for a future Nobel Prize. ‘They’ are induced pluripotent stem cells (or iPS for short). The discovery was that somatic cells from the adult body, whether from a hair, skin biopsy, cord-blood or even adipose tissue, can quite readily be changed back into pluripotent stem cells – ostensibly the state they were in shortly after conception – in the process, erasing the epigenetic modifications that make a brain cell different from, say, a liver cell.
This phenomenon is all the more remarkable because it is achieved by the introduction into cells of only three or four genes. The ease by which this process of cellular reprogramming can be achieved has turned dogma upside down and not surprisingly, iPS cells have captured the imagination of both researchers and the media alike. Why are they so important? Are they over-hyped? Here, I review the field and consider the impact this technique could have on our basic understanding of disease, the future of drug discovery and in regenerative medicine.
Cellular reprogramming
We all start out as one cell. Through a process of cell division and step wise specialisation, we end up as functional adults assembled from trillions of cells, of at least 250 different types. Each and every cell (with a few exceptions) has the same genetic make up and it is only through a series of epigenetic modifications that the combinations of genes specifying each cell type become differentially expressed. The current thinking is that this process of specialisation occurs stepwise and that cells pass from the so-called pluripotent state that characterises embryonic stem cells, through a series of intermediate states known as ‘progenitor cell’ populations that have limited capacity to self-renew and ultimately terminally differentiate to a single or a small number of mature cell types. Despite the pioneering work of John Gurdon in Xenopus laevis which showed reprogramming of adult nuclei to a totipotent state1, the assumption was that cellular differentiation occurred irreversibly in mammals. Twenty or so years later, cell fusion experiments showed that reactivation of embryonic gene expression could be achieved2-5. Then along came somatic cell nuclear transfer (SCNT) and Dolly the sheep6, thus demonstrating that reprogramming could occur in the right circumstances in most mammals too. Interestingly, SCNT so far has not been possible in the human, irrespective of any ethical con – siderations. Shinya Yamanaka extrapolated from the earlier observation that a single gene could convert adult fibroblasts into muscle cells and contemplated that introducing a number of genes into adult cells could convert them to an embryonic state. Drawing on the emerging knowledge of embryonic stem (ES) cells, they tested a battery of genes associated with the ES cell state and quite startlingly discovered that only three transcription factor genes (Oct3/4, Sox2, Klf4) and the oncogene c-Myc, were required to wind back the cellular clock and recreate pluripotent ES-like cells from mouse fibroblasts that could be differentiated into all three germ layers7, which were later shown to be germline competent8,9. Work from other groups confirmed and extended these findings into humans where the same four factors were found to be sufficient10-12 although one study showed that the pluripotency transcription factor Nanog and Lin28 (a miRNA processing enzyme) could also participate in driving reprogramming10. Subsequently, c-myc was shown not to be required in many circumstances11 and that some of the other factors were dispensable for reprogramming in certain cell types of a more embryonic nature. Moreover, Klf4 can be replaced in the process of reprogramming mouse embryonic fibroblasts by expressing the orphan nuclear receptor Esrrb13, and by members of the same protein family, Klf2 and Klf514. Equally, Sox2 can be replaced by Sox1 and Sox3, and c-myc by N-myc and L-myc14. Whether a particular factor is dispensable appears to be dependent on cell type. The only factor that remains essential and nonreplaceable by its closely related family members is Oct3/4 (a Pou-domain transcription factor also known as Pou5f1). However, recent evidence indicates that, at least in mouse cells, even the need to induce exogenous Oct3/4 expression can be circumvented by expressing the nuclear receptor Nr5a2 which triggers endogenous Oct3/4 expression15. This illustrates that the mere expression of these core transcription factors is enough to trigger a series of fortunate events which, in a small proportion of cells, leads to full reprogramming. Inter mediate states of partial reprogramming are also found to exist. Of great interest is the fact that miRNAs play an important role in controlling the transcriptional hierarchy required to maintain pluripotency16-18.
The many ways to make an iPS cell
The original iPS methods employed retroviruses to introduce the four cDNAs into cells7,8. Until recently, the majority of methods have relied on viral introduction of one form or another – a technique that often results in multiple random integration events, running the risk of insertion activation/inactivation which raises safety issues for downstream applications. In the process of reprogramming, the viral sequences are in – activated and the endogenous genes encoding the reprogramming factors reactivated but concerns surround the continued overexpression of pluripotency genes and the effects this has on a cell’s ability to be differentiated, or maintain that state in cell transplantation scenarios, risking the eventual formation of teratomas.
A significant technical development came recently with the effective introduction into somatic cells of a single vector expressing all four factors as a single cistron (either using a plasmid, lentivirus or transposon vectors) and then removal of all viral sequences using Cre-mediated excision19 or piggyBac transposition20,21. A further study used a non-integrating episomal vector to generate human iPS cells22, whilst another study used a non-replicating non-viral polycistronic vector to make mouse iPS cells23. Another useful concept was reported whereby four bicistronic lentiviruses (each reprogramming factor being used to a different fluorescent protein) were used to infect and monitor the reprogramming kinetics of somatic cells24.
In attempts to eliminate the use of viruses, other plasmid-based methods have been shown to work in mouse cells25,26 however, plasmid transfection of hESC is more problematic. A recent intriguing development was the demonstration that the proteins themselves could be introduced into cells via cellpenetrating peptides and this was found to be sufficient to enable reprogramming (albeit requiring multiple addition cycles and producing iPS colonies at low efficiency)27,28. Once optimised, this may be the method of choice in the future.
A more obvious, though perhaps more speculative, way to remove the requirement for viral based reprogramming is to employ small molecules. Small molecules have the advantage of being generally reversible, cheap and xenofree (hence GMP-compliant). Several reports have shown small molecules can improve the reprogramming process (reviewed29,30). Some of these, such as Valproic acid and TSA appear to work non-specifically by altering chromatin structure and reverse epigenetic modifications. Others appear to be targeting specific pathways governing pluripotency. Of particular interest is the recent demonstration that inhibition of the TGFβ pathway in mouse cells can replace the need for Sox231,32 by a mechanism thought to operate via the activation of Nanog. A recent study also provided evidence that a > 100-fold increase in reprogramming efficiency of human fibroblasts was observed by dual inhibition of TGFβ/activin signalling (using SB431542) and the MEK-ERK pathway (using PD-0325901)33. Since the signalling modules in mouse and human ES cells appear to be wired differently it will be of importance to determine whether separate toolkits of small molecules will be necessary for different species. It will be of considerable interest to search for small molecules that activate Oct3/4 expression since this event is the cornerstone of reprogramming. Hypoxic conditions, as found in the environment the early blastocyst is exposed to, have also recently been shown to improve reprogramming34. Vitamin C increases the reprogramming efficiency of iPS cells35 probably by alleviating cell senescence. Indeed, cell senescence and p53 status are thought to be a major roadblock to reprogramming36-41.
Which somatic cell types are the best sources to start with? In mice, iPS cells have been derived from embryonic and adult fibro – blasts, bone marrow cells, hepatocytes, gastric epithelial cells, pancreatic cells, neural stem cells and B lymphocytes. In humans, iPS cells have been derived from skin fibroblasts, keratinocytes, cord-blood cells and even less physiologically relevant cell lines like MRC5, and recently from surplus adipose cells. The number of examples is growing steadily. It is clear that the more terminally differentiated a cell is, the less prone it is to reprogramming42.
Patient contextualised cells
A key benefit of iPS cells is that from a single biopsy (which at least in principle can be as non-invasive as a hair plucked from a head), a virtually limitless supply of a variety of cell types can be produced. Because they are derived from a particular individual, in principle at least, they could be used to generate panels of patientspecific cells for various personalised medicine uses such as in vitro pre-treatment dosing studies, testing the effects of specific combinations of drugs and in predictive toxicology studies. Of great interest would be to make a library of different iPS lines from a wide range of the population of all ethnic groups. The logical conclusion of this ability is the possibility of generating cell products for repair (or someday even organs for transplantation?) that are entirely ‘self’ and hence remove the need to find matched donors. Whether the time and cost of making these cells and organs make it economically viable remains an unknown. Issues over ES/iPS-derived cell product safety remain and barriers generated by the need for case-bycase approval by the drug regulatory authorities are likely to be considerable. Nonetheless, only a few years ago, none of these possibilities were even conceivable, and given the amount of academic and commercial interest in the iPS field, improvements to the process are likely to be swift and significant.
Disease-contextualised cells
One difficulty hampering our understanding of disease states is the lack of physiological cell models. Animal models are often inadequate and sometimes highly misleading. Primary cells can sometimes be obtained but they are highly variable and difficult to propagate if the cell type of interest is anything other than a readily dividing cancer cell. Embryonic stem cells are of course able to be propagated easily, large numbers can be obtained in the laboratory, and with increasing ease they can be differentiated into many different cell types found in the body. The limitation is the fact that it is non-trivial to introduce defective genetic variants by homo – logous recombination (even by today’s standards) and then makes the assumption the precise nature of the genetic lesion is dominant and monogenic. Lesch-Nyhan diseasemimicking hESCs are one example of this approach43. An alternative way to generate disease models is to take cells from preimplantation genetic diagnosis (PGD) and derive stem cells from them. This approach has been used for cystic fibrosis44,45, Huntingdon’s disease44 and Fragile-X syndrome46. With the advent of iPS technology, patient biopsies can be used to generate iPS cells that can then be differentiated, at least in theory, into the cell type(s) affected in the disease state. Good examples of how this can be used to understand disease are in the case of Spinal Muscular Atrophy (SMA), an autosomal recessive disorder caused by mutations in the survival motor neuron (SMN) gene leading to motor neuron degeneration and progressive paralysis47. In another groundbreaking study, iPS cells were generated from an 82-year-old woman diagnosed with a familial form of amyotrophic lateral sclerosis (ALS)48. iPS cell technology has therefore opened the floodgates for disease research, with the list of disease-derived iPS cells increasing weekly from the early tour-de-force reports where iPS cells derived from nine different diseases were generated49 to more recent work that have generated Parkinson’s disease cells lacking viral reprogramming factors19. The excitement surrounding the use of iPS cell-technology is because for many other diseases, particularly those of the central nervous system, primary tissues are simply not available.
A future challenge will be to model complex polygenic diseases that display non-mendelian or low penetrance. Another major drawback with the study of disease in the culture dish might be encountered when studying long latency diseases such as Alzheimer’s or Parkinson’s diseases, however one can imagine strategies to ‘in vitro age’ cells by exposing them to the same environmental insults (for example UV or oxidative stress) that are thought to precipitate the disease in humans, the assumption being that these triggers are known. A third challenge is to differentiate the iPS cells with sufficient ease to the precise cell type involved in the disease state – to date only a handful of cell types can readily be produced in the laboratory. A fourth challenge relates to cell nonautonomous disease where one will have to go to the lengths of building 3D tissue models incorporating the diseased cells, in order to mimic the normal cellular milieu. A relatively simple known example of this scenario is the observed need to co-culture ALS motor neurons with glial cells in order to recapitulate the full disease pathology50,51.
Potential uses in drug discovery and regenerative medicine
The uses of iPS cells and their derivatives for the drug discovery industry are multi-fold. An immediate attraction is the reduced ethical and legal considerations, although if derived from patient material, consent is still required. Obvious uses lie in generating more physiologicallyrelevant cells for predictive toxicology (though these are also offered by using ES cells also) and potentially understanding how to stratify patients. The most exciting uses are in building new models for disease through the generation of cell types and screening systems hitherto unavailable.
Pluripotent cells might, in the future, allow cells or even tissues to be generated for repair. iPS technology means that there is the reasonable possibility of making cell products derived from a patient’s own cells, overcoming the rejection problem. Moreover, genetic correction of disease-specific iPS cells obtained from patients and then differentiating those corrected cells into a cell-based therapy product is an exciting if somewhat speculative propo – sition. Remarkably, this is not science fiction. Genetic correction of iPS cells using lentiviruses has been achieved from Fanconi Anaemia patient’s cells – with a time from biopsy to characterised, ‘corrected’ cell line in the order of four to five months52.
Give me a child until he is seven and I will give you the man
The explosion in the iPS research field is breathtaking. Human ES cells are a healthy 10 years old, but iPS cells are still in the scientific kindergarten at four. Much has to be learned about ES and iPS cells, from their basic biology and physiology to their utility in modelling diseases and creating cell types of choice. Differences do exist between ES and iPS cells and these will need to be thoroughly characterised. Reprogramming will need to be standardised, cell heterogeneity will need to be reduced and the propensity towards tumourigenesis tackled and eliminated. A fundamental understanding of how pluripotent cells work must underpin the commercial drive for therapy and product. Fortunately, the revolution in molecular and cellular biology over the last three decades has equipped researchers with ever more powerful ways of probing and scrutinising at the macro and micro scale. The life for iPS cells seems to be bright, at the very least interesting, and maybe rather rich.


Figure 1
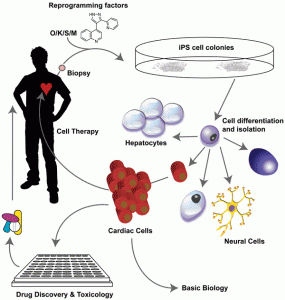

Figure 2
Acknowledgements
I would like to thank Professor Julie Frearson for guidance and support. ITI Lifesciences/Scottish Enterprise is acknowledged for financial support. Apologies for citations omitted due to space and time constraints.
References
- Gurdon, J.B., Adult frogs derived from the nuclei of single somatic cells. Dev Biol, 1962. 4: p. 256-73.
- Blau, H.M., C.P. Chiu, and C. Webster, Cyto plasmic activation of human nuclear genes in stable heterocaryons. Cell, 1983. 32(4): p. 1171-80.
- Tada, M., et al., Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr Biol, 2001. 11(19): p. 1553-8.
- Do, J.T. and H.R. Scholer, Nuclei of embryonic stem cells reprogram somatic cells. Stem Cells, 2004. 22(6): p. 941-9.
- Cowan, C.A., et al., Nuclear reprogramming of somatic cells after fusion with human embryonic stem cells. Science, 2005. 309(5739): p. 1369-73.
- Wilmut, I., et al., Viable offspring derived from fetal and adult mammalian cells. Nature, 1997. 385(6619): p. 810-3.
- Takahashi, K. and S. Yamanaka, Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell, 2006. 126(4): p. 663-76.
- Takahashi, K., et al., Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell, 2007. 131(5): p. 861-72.
- Okita, K., T. Ichisaka, and S. Yamanaka, Generation of germline-competent induced pluripotent stem cells. Nature, 2007. 448(7151): p. 313-7.
- Yu, J., et al., Induced pluripotent stem cell lines derived from human somatic cells. Science, 2007. 318(5858): p. 1917-20.
- Wernig, M., et al., c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell, 2008. 2(1): p. 10-2.
- Park, I.H., et al., Reprogramming of human somatic cells to pluripotency with defined factors. Nature, 2008. 451(7175): p. 141-6.
- Feng, B., et al., Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nat Cell Biol, 2009. 11(2): p. 197-203.
- Nakagawa, M., et al., Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol, 2008. 26(1): p. 101-6.
- Heng, J.-C.D., et al., The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell Stem Cell, 2010. 6(2): p. 167-74.
- Judson, R.L., et al., Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol, 2009. 27(5): p. 459-61.
- Gunaratne, P.H., Embryonic stem cell microRNAs: defining factors in induced pluripotent (iPS) and cancer (CSC) stem cells? Curr Stem Cell Res Ther, 2009. 4(3): p. 168-77.
- Viswanathan, S.R. and G.Q. Daley, Lin28: A microRNA regulator with a macro role. Cell, 2010. 140(4): p. 445-9.
- Soldner, F., et al., Parkinson’s disease patientderived induced pluripotent stem cells free of viral reprogramming factors. Cell, 2009. 136(5): p. 964-77.
- Kaji, K., et al., Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature, 2009. 458(7239): p. 771-5.
- Woltjen, K., et al., piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature, 2009. 458(7239): p. 766-70.
- Yu, J., et al., Human induced pluripotent stem cells free of vector and transgene sequences. Science, 2009. 324(5928): p. 797-801.
- Gonzalez, F., et al., Generation of mouse-induced pluripotent stem cells by transient expression of a single nonviral polycistronic vector. Proc Natl Acad Sci USA, 2009. 106(22): p. 8918-22.
- Papapetrou, E.P., et al., Stoichiometric and temporal requirements of Oct4, Sox2, Klf4, and c-Myc expression for efficient human iPSC induction and differentiation. Proc Natl Acad Sci USA, 2009. 106(31): p. 12759-64.
- Okita, K., et al., Generation of mouse induced pluripotent stem cells without viral vectors. Science, 2008. 322(5903): p. 949-53.
- Okita, K., et al., Generation of mouse-induced pluripotent stem cells with plasmid vectors. Nat Protoc, 2010. 5(3): p. 418-28.
- Zhou, H., et al., Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell, 2009. 4(5): p. 381-4.
- Kim, D., et al., Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell, 2009. 4(6): p. 472-6.
- Andrews, P.D. and J.A.F. Frearson, Influencing Stem Cell Fate with Small Molecules. European Pharmaceutical Review Digital, 2009. 1: p. 18-28.
- Feng, B., et al., Molecules that promote or enhance reprogramming of somatic cells to induced pluripotent stem cells. Cell Stem Cell, 2009. 4(4): p. 301-12.
- Ichida, J.K., et al., A small-molecule inhibitor of tgf-Beta signaling replaces sox2 in re programming by inducing nanog. Cell Stem Cell, 2009. 5(5): p. 491-503.
- Maherali, N. and K. Hochedlinger, Tgfbeta signal inhibition cooperates in the induction of iPSCs and replaces Sox2 and cMyc. Curr Biol, 2009. 19(20): p. 1718-23.
- Lin, T., et al., A chemical platform for improved induction of human iPSCs. Nat Meth, 2009. 6(11): p. 805-8.
- Yoshida, Y., et al., Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell, 2009. 5(3): p. 237-41.
- Esteban, M.A., et al., Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell, 2010. 6(1): p. 71-9.
- Utikal, J., et al., Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature, 2009. 460(7259): p. 1145-8.
- Li, H., et al., The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature, 2009. 460(7259): p. 1136-9.
- Marión, R.M., et al., A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature, 2009. 460(7259): p. 1149-53.
- Kawamura, T., et al., Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature, 2009. 460(7259): p. 1140-4.
- Banito, A., et al., Senescence impairs successful reprogramming to pluripotent stem cells. Genes & Development, 2009. 23(18): p. 2134-9.
- Hanna, J., et al., Direct cell reprogramming is a stochastic process amenable to acceleration. Nature, 2009. 462(7273): p. 595-601.
- Eminli, S., et al., Differentiation stage determines potential of hematopoietic cells for reprogramming into induced pluripotent stem cells. Nat Genet, 2009. 41(9): p. 968-76.
- Urbach, A., M. Schuldiner, and N. Benvenisty, Modeling for Lesch-Nyhan disease by gene targeting in human embryonic stem cells. Stem cells (Dayton, Ohio), 2004. 22(4): p. 635-41.
- Mateizel, I., et al., Derivation of human embryonic stem cell lines from embryos obtained after IVF and after PGD for monogenic disorders. Hum Reprod, 2006. 21(2): p. 503-11.
- Pickering, S.J., et al., Generation of a human embryonic stem cell line encoding the cystic fibrosis mutation deltaF508, using preimplantation genetic diagnosis. Reprod Biomed Online, 2005. 10(3): p. 390-7.
- Eiges, R., et al., Developmental study of fragile X syndrome using human embryonic stem cells derived from preimplantation genetically diagnosed embryos. Cell Stem Cell, 2007. 1(5): p. 568-77.
- Ebert, A.D., et al., Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature, 2009. 457(7227): p. 277-80.
- Dimos, J.T., et al., Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science, 2008. 321(5893): p. 1218-21.
- Park, I.H., et al., Disease-specific induced pluripotent stem cells. Cell, 2008. 134(5): p. 877-86.
- Di Giorgio, F.P. and K. Eggan, Non-cell autonomous effect of glia on motor neurons in an embryonic stem cell-based ALS model. 2007. 10(5): p. 608-14.
- Di Giorgio, F.P., Human embryonic stem cell-derived motor neurons are sensitive to the toxic effect of glial cells carrying an ALS-causing mutation. Cell Stem Cell, 2008. 3(6): p. 637-48.
- Raya, A., et al., A protocol describing the genetic correction of somatic human cells and subsequent generation of iPS cells. Nat Protoc, 2010. 5(4): p. 647-60
About the author
I obtained a Biochemistry B.Sc and Ph.D in Molecular Biology from the University of Sheffield. In 1993, I moved to the University of Dundee, to pursue my interest in signalling, first using budding yeast to understand Protein Phosphatase 1 function and then later using human cells characterised Aurora B protein kinase. Since 2007, I have been leading the Stem Cell Programme in the Drug Discovery Unit. I established the high-content screening capabilities and led several successful screening campaigns using human ES cells. Current interests lie in targeting signalling pathways using small molecules to: engineer cell fate; perform nuclear reprogramming and in the targeting of cancer stem cells.
email: [email protected]