Stathmin immobilisation on a chip using an oligo-cysteine tag
Posted: 27 March 2007 | | No comments yet
Protein chip technology is essential for high through-put functional proteomics. In this review the development of a novel protein tag consisting of five tandem cysteine repeats (Cys-tag) at C-terminus of proteins which was covalently attached to the surface of a maleimide-modified diamond-like carbon-coated silicon chip substrate is described.
Protein chip technology is essential for high through-put functional proteomics. In this review the development of a novel protein tag consisting of five tandem cysteine repeats (Cys-tag) at C-terminus of proteins which was covalently attached to the surface of a maleimide-modified diamond-like carbon-coated silicon chip substrate is described.
Protein chip technology is essential for high through-put functional proteomics. In this review the development of a novel protein tag consisting of five tandem cysteine repeats (Cys-tag) at C-terminus of proteins which was covalently attached to the surface of a maleimide-modified diamond-like carbon-coated silicon chip substrate is described.
Model proteins were created as a Cys-tagged enhanced green fluorescent protein (EGFP) and an EGFP-stathmin fusion protein which included an oligo-histidine tag at N-terminus to allow its purification from expressed proteins in Escherichia coli using Ni beads. The purified Cys-tagged proteins could be captured on the maleimide-coated chip substrate at efficiency that 50 pg of the fusion protein was detected by fluorescence and 5 pg (0.2 fmole) could be detected by combination with enhanced immunofluorescence. Furthermore, LC-MS/MS enabled us to identify protein candidates for binding partners of stathmin in mouse brain extracts on the chip. This highly sensitive immunodetection may be due to the strong covalent binding of proteins to the chip substrate via the Cys-tag combined with efficient exposure of the protein to interact with its binding partners in the surround solution. Thus, the Cys-tag should be useful for developing a novel protein printing method for protein chips that requires very low amounts of protein and can be used for high through-put analysis of protein-protein interactions.
The human genome project1,2 has opened biological and medical sciences focusing on proteome3,4. Following the development of DNA microarray chip technology for analysis of gene expression in genomic level, protein microarray chip technology for proteomic analysis has also advanced and been applied for the simultaneous identification, quantification and functional analysis of proteins5.
In early versions of protein chips, the chip surface was treated with an aldehyde-containing silane reagent to for Schiff’s base linkages to amines, lysine, or NH2-termini in proteins6. Nickel-coated slides have also been used to immobilise yeast proteome-wide recombinant proteins through histidine fusion tags (His-tags)7. Recently, nitrocellulose-coated glass slides have been employed for investigating protein-protein interactions8,9, finding specific antibodies from serum with antigen array10-11 and finding protein antigens with antibody arrays12-13. Bait proteins on chips must be firmly immobilised so that they remain attached after washing and incubation steps. The immobilisation must also allow interacting molecules to access and bind to the protein efficiently. In these regards, protein chips are not yet as advanced as DNA microarray chips and further modifications are needed to develop them into high-quality tools for proteomic analysis.
Here, a novel method for immobilising protein using a fusion tag of five tandem cysteines (Cys-tag) is described14. Thiol group of cysteine has been utilised for several protein analyses. For example, cysteine residues in any proteins could be labeled by maleimide group-linked fluorescent dyes15, applied for antibody-labeling or protein staining before or after electrophoresis in gel. Short tetracysteine motifs expressed inside of a recombinant protein could trap fluorophore later for the labeling of only the protein in vivo16,17 and in vitro17. For chip application, N-terminally cysteine-containing peptides were immobilsed on thioester slides18, which were applied to a study of kinase activity on peptide arrays19,20. The covalent binding property of cysteine is useful for the chip application of larger recombinant proteins via a fusion Cys-tag. A Cys-tag fusion of enhanced green fluorescent protein (EGFP)21 has been used for a marker of gene expression and protein targeting in intact cells and organisms because of its strong and stable green fluorescence. The purified Cys-tagged EGFP could be immobilised onto a maleimide-modified chip which was coated with diamond-like carbon and detected at femtomole levels by immunofluoresence and tandem mass spectrometry (LC-MS/MS)14. Furthermore, a Cys-tagged stathmin fused to EGFP was immobilised on the chip for a bait protein to analyse binding partners of stathmin by LC-MS/MS directly. This protein-tag and the related printing method described here allow the covalent binding of proteins to the surface of protein chips with high-performance for high through-put functional proteomics.
A novel technique of protein immobilisation has been developed for protein chips using a maleimide-modified silicon chip substrate as a solid support for covalently attaching proteins at their C-terminal thiol groups by introducing a Cys-tag (Figure 1). Since purification of target protein was essential for its binding to the substrate, a combination of a His-tag for purification with a Cys-tag for immobilisation is the most efficient for this system. The specific locations of the two tags in the recombinant protein may be important for expression in E. coli. A N-terminal His-tag and a C-terminal Cys-tag was the most efficient combination for the production, purification and immobilisation of the recombinant protein (Figure 2).Fluorescence measurement could detect a signal using 50 pg (0.5 µl spot of 100 pg /µl) of EGFP indicating that the protein’s three-dimensional conformation was essentially intact after immobilisation. Moreover, an immunological method using enhanced chemiluminescence could detect a signal when EGFP protein was used as little as 5 pg (0.2 fmol, 0.5 µl spot of 10 pg /µl). This is efficient when compared with other protein immobilisation systems, for example, coupling on nitrocellulose coated slides was accomplished with 1 µg/µl protein solution8, 0.1-12.8 ng/ml solution13, 10-50 ng/spot12, or coupling on BSA-N-hydroxysuccinimide slides via amino groups of 0.1- 0.5 µg/ml protein solution6, although Ni-coated slide immobilised with his-tagged 10-950 fg of protein/spot7 is more efficient. 3-5 mg/ml N-terminal cysteine containing peptides (usually CGG-and under 10 amino acids) solution were printed on thioestel slide19.
Target proteins are covalently immobilised to maleimide groups via a long linker and the EGFP fusion protein is attached in a uniform orientation and exposed well to the surrounding solution (Figure 3), also suggested in Ni-coated slides / His-tagged protein system7. This property allows efficient binding of the primary and secondary antibodies as well as the substrates for enhanced chemiluminescence. The main benefit of the maleimide/Cys-tag covalent binding over the Ni/His-tag affinity binding24 is stability of immobilisation theoretically, which should allow stringent washes and give lower background signals. Furthermore, chelating agents, which interfere with affinity of His-tagged proteins to Ni-coated slides, may not interfere with the immobilisation of Cys-tagged proteins to the maleimide chip substrate.
Nitrocellulose-coated glass slides have become popular as a substrate for protein immobilisation10-14. Proteins bind tightly to nitrocellulose via hydrophobic interactions25,26. Some highly charged proteins, however, may be difficult to bind to nitrocellulose and detergent prevents nitrocellulose from tight binding of proteins27. Also, nitrocellulose is expected to bind proteins in random orientations via internal hydrophobic residues. This may explain why nanogram quantities – a relatively large amount – of protein are utilised for nitrocellulose-coated chips. This system should also retain the three-dimensional structure of the protein, which will be required for the highly sensitive detection of biological functions on a chip, including protein-protein interactions and enzymatic activity. In the case of a fusion protein of Cys-tagged stathmin and EGFP, which is immobilised on the maelimde chip (stathmin chip), we were able to identify protein candidates for stathmin binding partners such as tublins, HSC70 family28 and other unreported proteins in brain extracts by LC-MS/MS (Figure 4) (manuscript in preparation). Furthermore, the stathmin chip may be useful for assay of site specific kinases29 because immunofluorescence and tandem mass spectrometry could identify phosphopeptides of stathmin on chip30 These findings will contribute to understanding the mechanisms for development and disorder of neuronal systems31.
In conclusion, a novel protein chip system, consisting of a maleimide-coated substrate and a fusion protein containing both Cys- and His-tags has been developed. This will be useful for the development of new protein chip applications32.
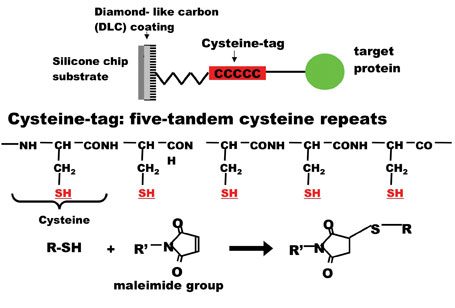

Figure 1: Covalent binding of Cys-tagged protein to maleimide group on diamond like carbon-coated chip substrate
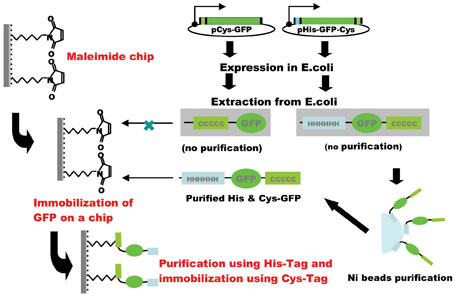

Figure 2: Construction of His- and Cys-tagged protein chip
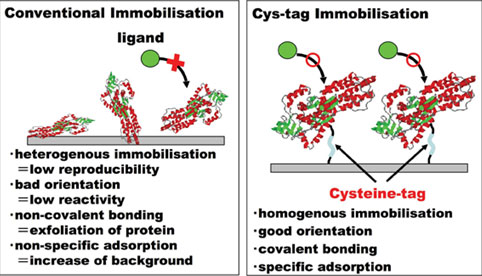

Figure 3: Advantages of Cys-tagged protein chip for functional proteomics
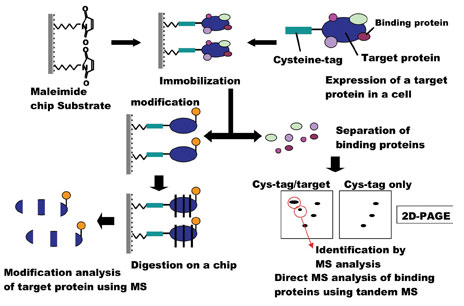

Figure 4: Identification of proteins on Cys-tagged protein chip by tandem mass spectrometry
Acknowledgements
I am grateful to my colleagues, Drs. Junko K. Akada, Yasuhiro Kuramitsu and Masanori Fujimoto in Yamaguchi University Graduate School of Medicine, Drs. Teruhisa Ichihara, Shuichi Kamei, Michifumi Tanga at Toyo Kohan Co., Ltd. Dr. Susumu Tsunasawa at Shimadzu Co., Ltd. and Dr. Andre Sobel. Plasmid construction works were carried out at the Gene Research Center of Yamaguchi University. This work was supported by grants-in-aid for science research 17590648 (to J.K.A.) and 16659142 (to K.N.) from the Ministry of Education, Science, Sports, and Culture of Japan.
References
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001, 409, 860-921.
- Venter, J. C.; Adams, M. D.; Myers, E. W.; Li, P. W.; Mural, R. J.; Sutton. G. G.; Smith, H. O.; Yandell, M., Evans, C. A. Evanc, C. A.; et. al. The Sequence of the Human Genome. Science. 2001, 291, 1304-1351.
- de Hoog, C. L.; Mann, M. Proteomics. Annu. Rev. Genomics Hum. Genet. 2004, 5, 267-293.
- Resing, K. A.; Ahn, N. G.Proteomic strategies for protein identification. FEBS Letters. 2005, 579, 885-889.
- Stoll, D.; Templin, M. F.; Bachmann, J.; Joos, T. O. Protein microarrays: Applications and future challenges. Current Opinion in Drug Discovery & Development.. 2005, 8, 239-252.
- MacBeath, G.; Schreiber, S.L. Printing proteins as microarrays for high-throughput function determination. Science, 2000, 289, 1790-1763.
- Zhu, H.; Bilgin, M.;Bangham, R.; Hall, D.; Casamayor, A.; Bertone, P.; Lan, N.; Jasen, R.; Bidlingmaier, S.; Houfek, T.; Mitchell, T.; Miller, P.; Dean, R. A.; Gerstein, M; Snyder, M. Global analysis of protein activities using proteome chips. Science, 2001, 293, 2101-2105.
- Espejo, A.; Cote, J.; Bednarek, A.; Richard, S.; Bedford, T. A protein-domain microarray identifies novel protein-protein interaction. Biochm. J. 2002, 367, 697-702.
- Yeast ProtoArrayTM PPI (Invitrogen) http://www.invitrogen.com
- Hueber, W.; Utz, P. J.; Steinman, L.; Robinson, W. H. Autoantibody profiling for the study and treatment of autoimmune disease. Arthritis Research 2002, 4, 290-295.
- Davies, D. H.; Liang, X.; Hernandez, J. E.; Randall, A.; Hirst, S.; Mu, Y.; Romero, K., M.; Nguyen, T. T.; Kalantari-Dehaghi, M., Crotty, S., Baldi, P.; Villarreal, L. P. Profiling the human immune response to infection by using proteome microarrays: High-throughput vaccine and diagnostic antigen discovery. Proc. Nat. Acad. Sci. 2005, 102, 547-552.
- Knezevic, V.; Leetahnakul, C.; Bichsel, V. E.; Worth, J. M., Prabha, V. V.; Gutkind, J. S.; Liotta, L. A.; Munson, P. J.; Petricoin III; E. F., Krizman, D. B. Proteimic profiling of the canner microenvironment by antibody arrays. Proteomics 2001, 1, 1271-1278.
- Michaud, G.; Salcius, M.; Zhou, F.; Bargham; R.; Bonin, J.; Guo, H.; Snyder, M.; Predki, P. F.; Schwetzer, B. I. Analysing antibody specificity with whole proteome microarrays. Nature Biotechnol. 2003, 21, 1509-1512.
- Ichihara, T.; Akada, J. K.; Kamei, S.; Ohsiro, S.; Sato, D.; Fujimoto, M.; Kuramitsu, Y.; Nakamura, K. A novel approach of protein immobilization for protein chips using an oligo-cysteine tag. J. Proteome Res. 2006, 5, 2144-2151.
- CyDye Maleimide Reactive Dye (GE helthcare) http:// www.gehealthcare.com
- Griffin, B. A.; Adams, S. R.; Tsien, R. Y. Specific covalent labeling of recombinant protein molecules inside live cells. Science 1998, 281, 269-272.
- Adams, S. R.; Campbell, R. E.; Gross, L. A.; Nartin, B. R.; Walkup, G. K.; Yao, Y.; Llopis, J.; Tsien, R. Y. New biarsenical ligands and tetracystein motifs for protein labeling in vitro and in vivo: Synthesis and biological applications. J. Am. Chem. Soc. 2002, 124, 6063-6076.
- Lesaichirre, M-L.; Uttamchandani, M.; Chen, G. Y. J; Yao, S. Q. Developing site-specific immobilisation strategies of peptides in microarray. Bioorg. Med. Chem. Lett. 2002, 12, 2079-2083.
- Uttamchandani, M.; Chen, G. Y. J; Lesaichirre, M-L.; Yao, S. Site-specific peptide immobilisation strategies for the rapid detection of kinase activity on microarrays. Methods in Molecular Biology, 2004, 264, 191-204.
- Zhu, H.; Klemic, J. F.; Chang, S.; Bertone, P.; Casamayor, A.; Klemic, K. G.; Smith, D.; Gerstein, M.; Reed, M. A.; Snyder, M.; Analysis of yeast protein kinases using protein chips. Nature Genet., 2000, 26, 283-289.
- Tsien, R. Y. The green fluorescent protein. Ann. Rev. Biochem. 1998, 67, 509-544.
- Staros, J. V. N-Hydroxysulfosuccinimide active esters: Bis (N-hydrosulfosuccinimide) esters of two diarboxylic acid are hydrophilic, membrane-impermeant, protein cross-linkers. Biochemistry 1982, 21, 3950-3955.
- Shimada, K.; Mitamura, K. Derivatisation of thil-containing compounds. J. Chromatogr. B. 1994, 659, 227-241.
- Terpe, K. Overview of tag protein fusions: from molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Biotechnol. 2003, 60, 523-533.
- Farrah, S. R.; Shah, D. O.; Ingram, O. Effects of chaotropic and antichaotropic agents on elution of poliovirus adsorbed on membrane filters. Proc. Natl. Acd. Sci. USA 1981, 78, 1229-1232.
- Gershoni, J.; Palade, G. E. Electrophoretic transfer of proteins from sodium dodecyl sulfate- polyacrylamide gels to positively charged membrane filter. Anal. Biochem. 1982, 124, 396-405.
- Nakamura, K.; Tanaka, T.; Kuwahara, A.; Takeo, K. Microassay for proteins on nitrocellose filter using protein dye-staining procedure. Anal. Biochem. 1985, 148, 311-319.
- Manceau, V.; Gavet, O.; Curmi, P; Sobel, A. Stathmin interaction with HSC70 family proteins. Electrophoresis, 1999, 20, 409-417.
- Curmi, P. A.; Gavet , O.; Charbaut, E.; Ozon, S.; Lachkar-Colmerauer, S.; Manceau, V.; Siavoshian, S.; Maucuer, A.; Sobel, A. Atahtmin and its phosphorylation family: General properties, biochemical and functional interaction with tubulin. Cell Structure and Function 1999, 24, 345-357.
- Nakamura, K.; Zhang, X.; Kuramitsu, Y.; Fujimoto, M.; Yuan, X.; Akada, J. K.; Aoshima-Okuda, M; Mitani, N.; Itoh, Y.; Katoh, T.; Morita, Y.; Nagasaka, Y.; Yamazaki, Y.; Kuriki, T.; Sobel, A. Analysis on heat stress-induced hyperphosphorylation of stathmin at serine 37 in Jurkat cells by means of two-dimensional gel electrophoresis and tandem mass spectrometry. J. Chromatogr. A. 2006, 1106, 181-189.
- Shumyatsky, G. P.; Malleret, G.; Shin, R-M.; Takizawa, S.; Tully, K.; Tsvetkov, E.; Zakharenko, S. S.; Joseph, J.; Vronskaya, S.; Yin, D.; Schubart, U. K.; Kandel, E. R.; Bolshakov, V. Y. stathmin, a Gene enriched in the amygdale, controls both learned and innate fear. Cell, 2005, 123, 697-709.
- Bertone, P.; Snyder, M.; Advances in functional protein microarray technology, FEBS Journal, 2005, 272, 5400-5411.




