How will MicroRNAs affect the drug discovery landscape?
Posted: 21 July 2007 | | No comments yet
The archetypal microRNAs, lin-4 and let-7, were discovered in the nematode worm Caenorhabditis elegans over a decade ago and, at that time, no one would have predicted that they would be anything other than an interesting feature of worm developmental biology. However, in recent years there has been an explosion of research activity in the field of microRNAs (miRNAs), so much so that the number of publications has almost doubled every year over the last five years (see Figure 1).
The archetypal microRNAs, lin-4 and let-7, were discovered in the nematode worm Caenorhabditis elegans over a decade ago and, at that time, no one would have predicted that they would be anything other than an interesting feature of worm developmental biology. However, in recent years there has been an explosion of research activity in the field of microRNAs (miRNAs), so much so that the number of publications has almost doubled every year over the last five years (see Figure 1).
The archetypal microRNAs, lin-4 and let-7, were discovered in the nematode worm Caenorhabditis elegans over a decade ago and, at that time, no one would have predicted that they would be anything other than an interesting feature of worm developmental biology. However, in recent years there has been an explosion of research activity in the field of microRNAs (miRNAs), so much so that the number of publications has almost doubled every year over the last five years (see Figure 1).
Fuelling this activity was the identification of miRNAs in many more organisms including humans, and especially the discovery that they are linked to human disease. In this article we review recent progress and developments that are advancing our understanding of the role of miRNAs in several human disease settings and therapeutic arenas, and how this may affect the drug discovery landscape.
MicroRNAs are evolutionarily conserved, noncoding RNA molecules, approximately 22 nucleotides in size, that negatively regulate target gene expression at the post-transcriptional level1-3. Originally ignored as that fraction of the genome colloquially termed “Junk DNA”, examples are now known in many species. Almost 40% of miRNAs are encoded in introns, 50% are intergenic and 10% are in exons. Release 9.2 (May 2007) of miRBase (the miRNA sequence database) http://microrna.sanger.ac.uk/4-6, contains 4584 entries representing hairpin precursor miRNAs, expressing 4430 mature miRNA products in primates, rodents, birds, fish, worms, flies, plants and viruses. The database contains 475 human miRNA sequences, just under half of which have been experimentally verified. The number of miRNA encoding genes is rapidly expanding, and this number may constitute between 1-4% of expressed genes in the genome7-9. Recent literature suggests that between 20-30% of all human genes may be subject to regulation by miRNAs and that each miRNA may, on average, contribute to the regulation of 200 or more mRNA targets10. Most miRNAs bind the 3’UTR of their target genes with imperfect sequence complementarities and repress translation initiation, though perfect complementarity can lead to mRNA cleavage in a process similar to that mediated in RNA interference (RNAi).
miRNAs in development and disease
The fact that many miRNAs are differentially expressed in tissues and developmental stages suggested their involvement in cell lineage determination and maintenance11. To date, the functional roles of only a relatively small number have been determined, however, this growing list includes several that are known to regulate important biological processes such as cell growth, differentiation and apoptosis12-16. Since these are all processes that can be dysregulated during disease, especially cancer, it is perhaps of little surprise that miRNAs became implicated. In fact, early clues for a role in cancer relevant processes came from studies of lin-4 and let-7 which were shown to have a role in the timing of key cellular events in C.elegans. Mutations in these genes were found to result in abnormalities in cell cycle and terminal differentiation which are hallmarks of cancer cells. Studies in which various miRNAs in the fruitfly, Drosophila melanogaster, have been either overexpressed or downregulated experimentally have also shown effects on cell proliferation and apoptosis17-19. Studies in mammalian systems have also shown key roles for miRNAs in development, by directing correct differentiation of cell lineages. Some examples include a role of miR-1 in heart development12, miR-181 in the differentiation of haematopoietic cells towards the B-cell lineage13, miR-196 in limb patterning14, miR-375 in pancreatic islet-cell development and regulation of insulin secretion20 and miR-143 during adipocyte differentiation15. The mammalian homologues of lin-4 and let-7 have been shown to control cell proliferation in human cell lines21,22 and are also associated with various cancers22-26. Malignant cell growth is characterised by loss of cellular identity, enhanced proliferation, and dysregulated cell death. Further functional analysis of identified and novel miRNAs is likely to reveal new connections between deregulation of proper cellular growth and differentiation and aberrant miRNA expression, providing further insight into the mechanisms that lead to tumourigenesis and other human diseases.
miRNAs and cancer
The first study to link miRNA expression and cancer resulted from the observation that the miR-15/miR-16 gene cluster is commonly deleted (>50% cases) in chronic lymphocytic leukaemia (CLL), and it was then confirmed that miR-15/miR-16 expression is frequently down regulated in CLL27,28. Subsequent studies have shown that many miRNAs are found at sites of chromosomal instability or genomic regions associated with cancer. For example, >90 miRNAs that map to the human genome are located in ‘fragile’ regions which are frequently amplified or deleted in tumours of breast, colon, lung, ovarian, gastric, liver, leukaemias and lymphomas29. More recently, expression ‘profiling’ of microRNAs has shown that tumours of different types have unique miRNA ‘signatures’, reflecting the developmental origin of that particular tumour30. The association of certain miRNAs and cancer has led to them being described collectively as ‘Oncogenic microRNAs’ or ‘Oncomirs’31.
In terms of the mechanisms involved, de-regulated expression of miRNAs in cancer can lead to the miRNA acting in one of two modes; in the first, by negatively regulating proto-oncogene expression, a miRNA can appear to act as a tumour suppressor. For example, by negatively regulating targets RAS or Bcl2, let-723 and miR-15a/miR-16-132,28 respectively, they begin to act as tumour suppressors. Conversely, by inhibiting targets that have tumour suppressor function (e.g., apoptotic genes, myc/myc pathway and E2F1), miR-21/Tropomyosin 133, miR-15534 or the miR-17-92 cluster35,36, are functioning as oncogenes (see Figure 2).
This is perhaps an oversimplification since it is possible that any given miRNA involved in cancer has a dual tumour suppressor/oncogene role that is dependent on a number of factors. These might include, for example, the cellular context and the balance of proliferative, apoptotic and differentiation activities created by the miRNAs and target mRNAs present within the cell and overlaid on other cellular regulatory systems.
miRNAs and inflammatory disease
A role for miRNAs has been implicated in inflammation, for example miR-155 has been shown to be involved in controlling inflammatory processes37,38, and miR-181a is an intrinsic modulator of T-cell sensitivity and selection39.
O’Connell et al noted that the mammalian inflammatory response to infection involves the induction of several hundred genes, a process that must be carefully regulated to achieve pathogen clearance and prevent the consequences of unregulated expression, such as cancer. As previously described, miRNAs have emerged as a class of gene expression regulators that have also been linked to cancer. However, the relationship between inflammation, innate immunity, and miRNA expression is just beginning to be explored. O’Connell et al used microarray technology to identify miRNAs induced in primary murine macrophages after exposure to polyriboinosinic:polyribocytidylic acid or the cytokine IFN-b. miR-155 was the only miRNA of those tested that was substantially up-regulated by both stimuli. It was also induced by several Toll-like receptor ligands through myeloid differentiation factor 88- or TRIF-dependent pathways, whereas up-regulation by IFNs was shown to involve TNFa autocrine signalling. Pharmacological inhibition of the kinase JNK blocked induction of miR-155 in response to either polyriboinosinic:polyribocytidylic acid or TNFa, suggesting that miR-155- inducing signals use the JNK pathway. Together, these findings characterised miR-155 as a common target of a broad range of inflammatory mediators. Importantly, because miR-155 is known to function as an oncogene, these observations identified a potential link between inflammation and cancer.
Another example comes from Rodriguez et al38 where they have shown that bic/miR-155 is required for the function of B and T lymphocytes and dendritic cells. Transcriptome analysis of bic/miR-155–deficient CD4+ T-cells identified a wide spectrum of miR-155-regulated genes, including cytokines, chemokines, and transcription factors. They also showed that mice that are deficient for bic/miR-155, demonstrating increased lung airway remodelling.
With regards to miR-181a and T-cells, Li et al39 used an antagomir (i.e. an antisense oligonucleotide that silences miRNA expression in vivo) to reduce miR-181a expression. miR-181a function was tested in DP (CD4+/CD8+ double positive thymocytes) cell selection using foetal thymic organ culture (FTOC) with or without antagomir treatment. 5C.C7 TCR transgenic mice were crossed onto an invariant chain knockout (Ii–/–) background to arrest thymocytes at the DP stage. Presenting exogenous 5C.C7 TCR antigens, such as agonist (MCC, Moth Cytochrome C), weak agonist (MCC 102S), or antagonist (MCC 99R), induced efficient negative selection within 48 hours, and introducing antagomir-181a to these cultures resulted in a greater than 50% inhibition.
miRNAs in metabolic, neurological, and other disease settings
A number of roles that hint at their likely importance in diseases other than cancer and inflammation have been uncovered for various miRNAs in mammals. For example, miR-375, a pancreatic islet specific miRNA cloned from mouse, was shown to regulate glucose dependent insulin secretion through its target, myotrophin. adenoviral over-expression of miR-375 suppressed glucose stimulated insulin secretion in the murine insulinoma cell line MIN6, whilst siRNA mediated suppression of miR-375 enhanced insulin secretion20. In this study, myotrophin (Mtpn) is a target of miR-375, and siRNA-mediated knockdown of Mtpn was shown to have the same phenotype as knockdown of the miRNA (i.e. stimulation of insulin secretion and exocytosis). Functional genomic approaches, such as the use of antisense oligonucleotides (ASOs) and microarray analysis of gene expression, have helped to uncover a role for miR-122, a liver specific miRNA, in lipid metabolism40,41. ASO inhibition of miR-122 increases fatty acid oxidation rate at the same time as decreasing fatty acid and sterol synthesis rates41. This in turn resulted in reduced plasma cholesterol and triglyceride levels in normal mice and, in an obese mouse model (ob/ob) resulted in reduced liver steatosis.
A number of miRNAs have been shown to be brain specific42,43, with important roles in brain development44,45. For example miR-9 appears to be prominently expressed early during development in proliferating neuronal precursors44,45. miR-124 however appears to be more widely expressed in differentiated neurones44-46. Given this role in normal neuronal growth and development it is likely therefore that aberrant expression and/or functional mutations in miRNAs will be important in diseases of the CNS. Indeed, a few recent examples have emerged to suggest a link between miRNAs and neurological disorders. For example, the SLITRK1 gene which is associated with Tourette’s syndrome, has been found to carry a mutation in its 3’ UTR that enhances repression of SLITKR1 mRNA by miR-189, possibly by preventing promotion of dendritic growth47. Another miRNA, miR-134, has also recently been implicated in development of dendritic spines48.
miRNAs as diagnostic and prognostic tools
The consensus emerging from a number of studies in which the expression profiles of miRNAs have been examined is that altered miRNA expression is commonly associated with cancer30,49. Various platforms (microarray, QPCR) have been employed in the generation of ‘global’ miRNA expression ‘signatures’30,44,45,50,51 that are being used to classify various cancers and to define useful prognostic biomarkers22,24,25,30,32. To date, a growing number of studies have compared miRNA expression profiles across tumours derived from different tissue types. For example, in a landmark study, Lu et al found that miRNA expression is primarily downregulated in tumour cells and is also a powerful predictor of tumour origin and differentiation state, reflecting that tumour’s developmental history30. Further efforts to profile miRNA expression across various tumours have also identified differentially expressed miRNAs in colon cancer49,52,53, glioblastoma54,55, lung49,52,56,57, lymphoma24,32 and testicular germ cell cancers58. Taken together, these observations underscore the diagnostic potential of miRNAs in cancer.
Downregulation of let-7 miRNA expression has been reported in lung cancer and this has been associated with poor prognosis22,23. For example, a study that looked at the correlation between let-7 expression level and post operative survival rates in 143 human lung cancer patients five years after surgery, showed that reduced expression was associated with poor prognosis22. Reduced let-7 expression was also observed in 60% of cancer cell lines examined and also in 44% of lung tumours, compared to normal adjacent tissue. Patients with squamous cell carcinoma of the lung (compared to normal) had decreased levels of let-7 expression which is associated with high levels of RAS. Finally, various let-7 family members have been mapped to deleted regions of the genome in lung cancer29. This information, taken together with data from various functional studies, suggests that let-7 is diagnostic for patients with non-small cell lung carcinoma (NSCLC) and prognostic for their post operative survival22,23,56,59. Indeed, let-7 expression may be unique for lung cancer progression since although markedly reduced in this form of cancer, it has not been found to be consistently decreased in other tumour types.
Therapeutic intervention of miRNAs
As described above, miRNAs play a pivotal role in many cellular and disease processes and, therefore, their expression could potentially be modulated in order to lead to therapeutic benefit. There are two routes by which this could be achieved: augmentation of miRNA activity or inhibition of miRNA activity.
Overexpression of miRNAs can be achieved by either using double-stranded miRNA mimetics, or by using an expression vector (in a similar manner to gene therapy). miRNA mimetics would have a similar structure to small interfering RNAs (siRNAs), and could benefit from all the recent progress with siRNA therapeutics e.g. oligonucleotide chemistries and formulations. However, they would also have to cross the same hurdles as therapeutic siRNAs e.g. delivery at therapeutic (but not toxic) doses to the relevant tissues. At the moment there is very little miRNA-specific data for this approach. Similarly, short hairpin RNA (shRNA)-based expression vectors could pave the way for the future development of miRNA gene therapy. Plasmid vectors and viral vectors are both being investigated by many groups for the delivery of RNAi effector molecules. For example, Nucleonics Inc. now have FDA approval to commence a Phase I clinical trial using a plasmid that expresses four different shRNA sequences that target human hepatitis B virus. Alternatively, adenovirus and adeno-associated virus (AAV) vectors have been used to deliver shRNAs to the liver60 and the brain61.
Inhibition of miRNA expression and/or activity has been a more extensively used route to understand miRNA function. Antisense oligonucleotides have been generated against a number of miRNAs in order to ‘knockdown’ the expression of the miRNA, and these ‘antagomirs’ have been shown to dramatically affect the phenotype of cellular processes in vitro. As described above, antagomirs directed against endogenous miR-181a have shown that this miRNA is an intrinsic modulator of T-cell sensitivity and selection39. Encouragingly, antagomirs have also been used successfully in vivo. Intravenous delivery of antagomirs against miR-16, miR-122, miR-192 and miR-194 resulted in a marked reduction of corresponding miRNA levels in liver, kidney, heart, intestine, fat, skin, bone marrow, muscle, ovaries and adrenal glands40. Further analysis showed that the 3’ untranslated regions of upregulated genes were strongly enriched in miR-122 recognition motifs, whereas downregulated genes were depleted in theses motifs. Cholesterol biosynthesis was predicted to be affected by miR-122, and plasma cholesterol measurements revealed reduced levels in antagomir-122-treated mice. These studies were extended by Esau et al41 who showed that knockdown of miR-122 in a diet-induced obesity mouse model resulted in decreased plasma cholesterol levels and a significant improvement in liver steatosis (see Figure 3).
Summary and future perspectives
In this article, we have briefly reviewed recent progress and developments that are advancing our understanding of the role of miRNAs in human disease, and how this may affect the drug discovery landscape. Given the pace at which research in this area is advancing, we anticipate that rapid progress in the next few years will lead to the entire ‘miRNome’ becoming known, and all of its members being cloned and sequenced. Coupled with this will be a better understanding of miRNA targets, and miRNA target site interactions. It is likely that patterns of miRNA expression will be used in a similar manner to that presently used in transcriptomic analyses. For example, miRNAs could be used routinely as either diagnostic and/or prognostic biomarkers (e.g., for cancer and other diseases), and this may lead to novel therapeutic targets and pathways, for example, through the identification of miRNAs that regulate specific cellular targets or pathways as in the case of myotrophin/insulin secretion. miRNAs may become therapeutic targets in their own right (e.g., miR-375 in the regulation of insulin metabolism) using siRNA, shRNA, antisense and/or other modified oligonucleotides. Finally, progress in the understanding of miRNAs may lead to the use of miRNAs as therapeutics.
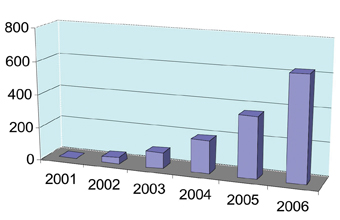

Figure1: The number of microRNA publications has nearly doubled each year since 2001
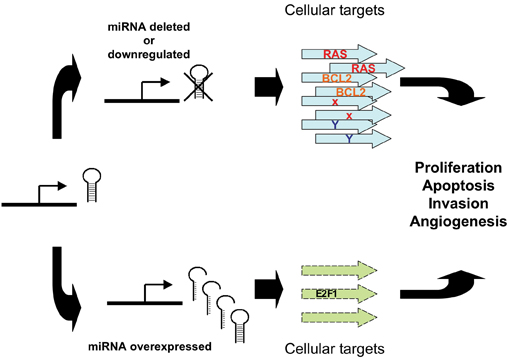

Figure 2: Abnormal miRNA expression can lead to tumourigenesis. Reduced expression of a mirRNA that acts as a tumour suppressor can lead to tumour formation. Conversely, overexpression of an oncogenic miRNA can contribute to tumour formation through increased inhibition of tumour suppressor Genes or other target genes that regulate growth or apoptosis.
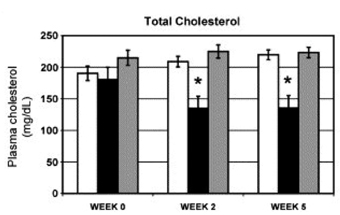

Figure 3: Cholesterol lowering in diet-induced obesity mouse model after miR-122 inhibition. C57Bl/6 mice that had been fed a high-fat diet for 19 weeks were treated subcutaneously with 12.5 mg/kg miR-122 or control ASO twice weekly for 5 1/2 weeks. n = 5. Plasma cholesterol levels at various time points after start of treatment. Error bars = SEM. *p < 0.05. White bar = saline, black bar = miR-122 antisense oligonucleotide, grey bar = control oligonucleotide. From Esau et al,: miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metabolism 2006 3, 87-98.
References
- Kim, V.N., MicroRNA biogenesis: coordinated cropping and dicing. Nature Rev. Mol.Cell Biol. 2005; 6, 376-385.
- Ambros, V., MicroRNAs: tiny regulators with great potential. Cell 2001; 107(7), 823-826.
- Chendrimada, T.P., Finn, K.J., Ji, X., Baillat, D., GregoryR.I., Liebhaber, S. A., Pasquinelli, A.E. and Shiekhattar, R. microRNA silencing through RISC recruitment of eIF6 Nature 2007; 447 (7146), 823-829.
- Griffiths-Jones S. The miRBase Database: microRNA sequences, targets and nomenclature, in microRNAs: Biology, Function and Expression, Clarke, N.J and Sanseau, P, Editors 2007, DNA Press: Eagleville, PA.p. 323-334.
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res., 2006, 34, D140-D144.
- Griffiths-Jones S.The miRNA Registry. Nucleic Acids Res., 2004, 32, D109-D111.
- Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP: Vertebrate MicroRNA Genes. Science 2003, 299:1540.
- Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, Sharon E, Spector Y, Bentwich Z: Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet 2005, 37:766-770.
- Berezikov E, Guryev V, van de BJ, Wienholds E, Plasterk RH, Cuppen E: Phylogenetic shadowing and computational identification of human microRNA genes. Cell 2005, 120:21-24.
- Lim, L.P, Lau, N.C, Garrett-Engele, P, Grimson, A, Schelter, J.M, Castle, J, Bartel, D.P, Linsley, P.S, Johnson, J.M: Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005, 433;769-773.
- Wienholds, E., Plasterk, R.H: microRNA function in animal development. Febs Letters 2005, 579: 5911-5922.
- Zhao Y, Samal E, Srivastava D: Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 2005, 436:214-220.
- Chen CZ, Li L, Lodish HF, Bartel DP: MicroRNAs Modulate Hematopoietic Lineage Differentiation. Science 2004, 303:83-86.
- Hornstein E, Mansfield JH, Yekta S, Hu JK-H, Harfe BD, McManus MT, Baskerville S, Bartel DP, Tabin CJ: The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature 2005, 438:671-674.
- Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV, Sun Y, Koo S, Perera RJ, Jain R, Dean NM, Freier SM, Bennett CF, Lollo B, Griffey R: MicroRNA-143 regulates adipocyte differentiation. J.Biol.Chem. 2004, 279:52361-52365.
- Xu P, Vernooy SY, Guo M, Hay BA: The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Current Biology 2003, 13:790-795.
- Hipfner DR, Weigmann K, Cohen SM: The bantam Gene Regulates Drosophila Growth. Genetics 2002, 161:1527-1537.
- Leaman D, Chen PY, Fak J, Yalcin A, Pearce M, Unnerstall U, Marks DS, Sander C, Tuschl T, Gaul U: Antisense-Mediated Depletion Reveals Essential and Specific Functions of MicroRNAs in Drosophila Development. Cell 2005, 121:1097-1108.
- Nairz K, Rottig C, Rintelen F, Zdobnov E, Moser M, Hafen E: Overgrowth caused by misexpression of a microRNA with dispensable wild-type function. Developmental Biology 2006, 291:314-324.
- Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M: A pancreatic islet-specific microRNA regulates insulin secretion. Nature 2004, 432:226-230.
- Lee YS, Kim HK, Chung S, Kim KS, Dutta A: Depletion of Human Micro-RNA miR-125b Reveals That It Is Critical for the Proliferation of Differentiated Cells but Not for the Down-regulation of Putative Targets during Differentiation. J.Biol.Chem. 2005, 280:16635-16641.
- Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, Mitsudomi T, Takahashi T: Reduced Expression of the let-7 MicroRNAs in Human Lung Cancers in Association with Shortened Postoperative Survival. Cancer Res 2004, 64:3753-3756.
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ: RAS is regulated by the let-7 microRNA family. Cell 2005, 120:635-647.
- Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M, Dell’Aquila ML, Alder H, Rassenti L, Kipps TJ, Bullrich F, Negrini M, Croce CM: MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc. Natl. Acad. Sci. 2004, 101:11755-11760.
- Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Menard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM: MicroRNA gene expression deregulation in human breast cancer. Cancer Res 2005, 65:7065-7070.
- Sonoki T, Iwanaga E, Mitsuya H, Asou N: Insertion of microRNA-125b-1, a human homologue of lin-4, into a rearranged immunoglobulin heavy chain gene locus in a patient with precursor B-cell acute lymphoblastic leukemia. Leukemia 2005, 19(11):2009-2010.
- Calin G.A, Dumitru C.D, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce C.M: Frequent deletions and down-regulation of microRNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukaemia. Proc.Natl. Acad.Sci. 2002, 99: 15524-15529.
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu Cg, Kipps TJ, Negrini M, Croce CM: miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc.Natl. Acad.Sci. 2005, 102:13944-13949.
- Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM: Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. 2004, 101:2999-3004.
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR: MicroRNA expression profiles classify human cancers. Nature 2005, 435:834-838.
- Esquela-Kerscher, A, Slack, F.J: Oncomirs-microRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6:259-269.
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, Iuliano R, Palumbo T, Pichiorri F, Roldo C, Garzon R, Sevignani C, Rassenti L, Alder H, Volinia S, Liu Cg, Kipps TJ, Negrini M, Croce CM: A MicroRNA Signature Associated with Prognosis and Progression in Chronic Lymphocytic Leukemia. N Engl J Med 2005, 353:1793-1801.
- Zhu, S, Si, M.L, Wu, H, Mo, Y.Y. microRNA-21 targets the tumor suppressor gene tropomyosin1 (TPM1). J. Biol. Chem 2007, 282(19): 14328-14336.
- Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE: Accumulation of miR-155 and BIC RNA in human B cell lymphomas. PNAS 2005, 102:3627-3632.
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM: A microRNA polycistron as a potential human oncogene. Nature 2005, 435:828-833.
- O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT: c-Myc-regulated microRNAs modulate E2F1 expression. Nature 2005, 435:839-843.
- O’Connell, R.M., Taganov, K.D., Boldin, M.P., Cheng, G., Baltimore, D.: MicroRNA-155 is induced during the macrophage inflammatory response. PNAS 2007, 104(5), 1604-1609.
- Rodriguez, A., Vigorito, E., Clare, S., Warren, M.V., Couttet, P., Soond, D.R., van Dongen, S., Grocock, R.J., Das, P.P., Miska, E.A., Vetrie, D., Okkenhaug, K., Enright, A.J., Dougan, G., Turner, M., Bradley, A.: Requirement of bic/microRNA-155 for Normal Immune Function. Science 2007, 316, 608-611.
- Li,Q-J., Chau, J., Ebert, P.J.R., Sylvester, G., Min, H., Liu, G., Braich, R., Manoharan, M., Soutschek, J., Skare, P., Klein, L.O., Davis, M.M., Chen, C.-Z.: miR-181a Is an Intrinsic Modulator of T-cell Sensitivity and Selection. Cell 2007, 129, 147-161.
- Krutzfeldt, J, Rajewsky, N, Braich, R, Rajeev, K.G, Tuschl, T, Manoharan, M, Stoffel, M.: Silencing of microRNAs in vivo with ‘antagomirs’. Nature 2005, 438 (7068): 685-689.
- Esau, C, Davis, S, Murray, S.F, Yu, X.X, Pandey, S.K, Pear, M, Watts, L, Booten, S.L, Graham, M, McKay, R, Subramaniam, A, Propp, S, Lollo, B.A, Freier, S, Bennett, C.F, Bhanot,S, Monia, B.P.: miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006, 3 (2): 87-98.
- Lagos-Quintana, M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W., Tuschl, T.: Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002, 12 (9), 735-739.
- Kim, J., Krichevsky, A., Grad, Y., Hayes, G.D., Kosik, K.S., Church, G.M., Ruvkun, G.: Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc. Natl. Acad. Sci. U. S. A., 2004, 101 (1), 360-365.
- Miska, E.A., Alvarez-Saavedra, E., Townsend, M., Yoshii, A., Sestan, N., Rakic, P., Constantine-Paton, M., Horitz, H.R.: Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004, 5 (9) R68.
- Krichevsky, A.M., King, K.S., Donahue, C.P., Khrapko, K., Kosik, K.S.: A microRNA array reveals extensive regulation of microRNAs during brain development. RNA 2003, 9 (10) 1274-1281.
- Deo, M., Yu, J.Y., Chung, K.H., Tippens, M., Turner, D.L.: Detection of mammalian microRNA expression by in situ hybridisation with RNA oligonucleotides. Dev. Dyn. 2006, 235 (9) 2538-2548.
- Abelson, J.F., Kwan, K.Y., O’Roak, B.J., Baek, D.Y., Stillman, A.A., Morgan, T.M., Mathews, C.A., Pauls, D.L., Rasin, M.R., Gunel, M., Davis, N.R., Ercan-Sencicek, A.G., Guez, D.H., Spertus, J.A., Leckman, J.F., Dure, L.S.4th, Kurlan, R., Singer, H.S., Gilbert, D.L., Farhi, A., Louvi, A., Lifton, R.P., Sestan, N., State, M.W.: Sequence variants in SLITRK1 are associated with Tourette’s syndrome. Science 2005, 310 (5746) 317-320.
- Schratt, G.M., Tuebing, F,. Nigh, E.A., Kane, C.G., Sabatini, M.E., Kiebler, M., Greenberg, M.E.: A brain-specific microRNA regulates dendritic spine development. Nature 2006, 439 (7074) 283-289.
- Volinia S, Calin GA, Liu Cg, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce, CM: A microRNA expression signature of human solid tumors defines cancer gene targets. Proc.Natl.Acad.Sci. 2006, 103:2257-2261.
- Thomson JM, Parker J, Perou CM, Hammond SM: A custom microarray platform for analysis of microRNA gene expression. Nat Meth 2004, 1:47-53.
- Nelson PT, Baldwin DA, Scearce LM, Oberholtzer JC, Tobias JW, Mourelatos Z: Microarray-based, high-throughput gene expression profiling of microRNAs. Nature Methods 2004, 1:155-161.
- Jiang, J, Lee, E.J, Gusev, Y and Schmittgen, T.D. Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res. 2005, 33(17): 5394-5403.
- Michael, M.Z, OCSM, Van Holst Pellekaan, N.G, Young, G.P and James, R.J. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol. Cancer. Res. 2003, 1(12): 882-891.
- Chan, J.A, Krichevsky, A.M and Kosik, K.S. MicroRNA-21 is an antiapototic factor in human glioblastoma cells. Cancer Res. 2005, 65(14):6029-6033.
- Ciafre, S.A, Galardi, S, Mangiola, A, Ferracin, M, Liu, C.G, Sabatino, G, Negrini, M, Maira, G, Croce, C.M and Farace, M.G. Extensive modulation of a set of microRNAs in primary gliobalstoma. Biochem. Biophys. Res. Commun. 2005, 334(4): 1351-1358.
- Yanaihara N, Caplen N, Bowman E, Seike M, Kumanoto K, Yi M, Stephens R, Okamoto A, Yokota J, Tanaka T, Calin G, Liu C, Croce C, Harris C: Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 2006, 9:189-198.
- Hayashita, Y, Osada, H, Tatematsu, Y, Yamada, H, Yanagisawa, K, Tomida, S, Yatabe, Y, Kawahara, K, Sekido, Y, Takahashi, T. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res., 2005, 65(21): 9628-9632
- Voorhoeve, P.M, le Sage, C, Schrier, M, Gillis, A.J, Stoop, H, Nagel, R, Liu, Y.P, van Duijse, J, Drost, J, Griekspoor, A, Zlotorynski, E, Yabuta, N, De Vita, G, Nojima, H, Looijenga, L.H and Agami, R. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell, 2006, 124(6) : 1169-1181
- Grosshans H, Johnson T, Reinert KL, Gerstein M, Slack FJ: The Temporal Patterning MicroRNA let-7 Regulates Several Transcription Factors at the Larval to Adult Transition in C. elegans. Developmental Cell 2005, 8:321-330.
- Uprichard, S.L., Boyd, B., Althage, A., Chisari, F.V.: Clearance of hepatitis B virus from the liver of transgenic mice by short hairpin RNAs. PNAS 2005, 102(3), 773-778.
- Xia, H., Mao, Q., Paulson, H.L., Davidson, B.L.: siRNA-mediated gene silencing in vitro and in vivo. Nat. Biotechnol. 2002, 20(10), 1006-1010.
Neil J. Clarke
Neil J. Clarke is a molecular cell biologist and scientific manager in the Biopharmaceutical Discovery Technology Group at GlaxoSmithKline, Stevenage, UK. He has extensive functional genomics expertise including transgenics and RNAi and is co-editor of a recently published book on the biology, function and expression of miRNAs.
Mark R. Edbrooke
Mark R. Edbrooke is a molecular cell biologist and scientific manager in the Respiratory and Inflammation Discovery Technology Group at GlaxoSmithKline, Stevenage, UK. He has extensive functional genomics expertise including RNAi, antisense, and other knockdown technologies.




