A bright future for drug discovery and development
Posted: 11 November 2005 | | No comments yet
High content imaging (HCI), the combination of automated fluorescence microscopy with quantitative image analysis, has been opening new dimensions in cytometry. This article gives an overview on the growing spectrum of applications and an outlook on the future use of this still rapidly developing technology.
High content imaging (HCI), the combination of automated fluorescence microscopy with quantitative image analysis, has been opening new dimensions in cytometry. This article gives an overview on the growing spectrum of applications and an outlook on the future use of this still rapidly developing technology.
High content imaging (HCI), the combination of automated fluorescence microscopy with quantitative image analysis, has been opening new dimensions in cytometry. This article gives an overview on the growing spectrum of applications and an outlook on the future use of this still rapidly developing technology.
The pharmaceutical industry is challenged by apparently opposing goals: better attrition rates should be achieved together with faster introduction of new drugs into the market. As a consequence, the integration of new technologies into drug discovery and development is crucial to obtain better data for decision making. New tools for the analysis of gene and protein expression patterns are currently changing drug development. However, cells – not genes – are the smallest functional units of life and the need is increasing to improve throughput and information content of functional assays. Several methods have been developed that extract deeper and broader information from functional assays, including microscope based methods, complex multiparameter flow cytometry and techniques analysing metabolic patterns. The term ‘high content analysis’ is used for a number of methods delivering multiplexed readouts, but the present review is focused on high content imaging (HCI). This technology, termed ‘imaging cytometry’, ‘subcellular imaging’ or ‘automated quantitative fluorescence microscopy’ is based on fluorescently stained cell samples that are visualised with an automated fluorescence microscope or confocal microscope system. The images are automatically analysed by specific image analysis tools. In other words, HCI is combining fluorescence microscopy, automated data acquisition and image analysis. Fluorescence microscopy is a mature technique – for decades cell biologists or pathologists have used microscopy based assays for mechanistic and functional analyses. With automatic image capturing and analysis the enormous amount of information from these images becomes fully accessible. Automation allows the use of fluorescence microscope based assays with virtually any throughput needed, while the image analysis software can deliver unbiased quantification of any visible property of an image. Analysis algorithms identify single cells, commonly via nuclear staining, and quantify all image parameters on the single cell level (imaging cytometry), including parameters that could hardly be measured by an experienced scientist using the microscope.
The first commercial HCI systems became available in the mid 1990s. Currently more than a dozen such instruments are on the market and great progress has been made with image acquisition hardware, data handling and image analysis solutions. The systems are still developing rapidly and further steps are yet needed to enable HCI analysis to become still more flexible and easier to use. Nevertheless, an impressive array of different HCI applications has already been published. This review aims to give an overview on the application spectrum for HCI analysis of cellular function. Applications cover primary and secondary drug screening; safety related profiling; target validation; mechanistic investigations in pharmacology and toxicology and even biomarker analysis in clinical samples. Although HCI was often perceived as merely a tool for functional screening, microscope based HCI opens up unique opportunities for functional assays that reach far beyond screening.
A picture tells more than 1000 words
Microscope based functional assays have been applied for a long time. However, the enormous costs of having highly trained specialists spending weeks behind a microscope limited the use of this functional readout in the past to indispensable regulatory tests or to assays in very small scale. By means of automatisation, HCI analysis gives microscope based analysis sufficient throughput to consider the broad use of microscope based assays as functional assays.
An example for an authority accepted microscope based test is the micronucleus assay, applied for the prediction of genotoxicity. Very promising approaches have begun to analyse this endpoint on HCI systems. Validation of the HCI assay as an authority accepted test has not yet started, but a number of pharma companies are evaluating the application of this HCI assay as a genotoxicity screening tool in research settings.
For functional assays image based methods offer clear advantages when localisation, colocalisation, translocation, movement or outgrowth are measured. Signal transduction pathways involve translocation events and, for many drugable targets, activation status and sub-cellular localisation are functionally linked. Most G-protein coupled receptors (GPCR), a big group of potential targets, are desensitised after activation by arrestin binding and internalisation. Different HCI assays for interference with GPCR signalling have been described, that analyse GPCR activation by localising a GPCR GFP fusion protein1,2 or an arrestin GFP fusion protein3. Both assay types have been successfully applied to drug screening. For the analysis of EGF receptor function another strategy has been described; internalisation of this surface receptor was quantified via uptake of labelled epidermal growth factor4.
Translocation into the nuclear compartment is easily measurable by HCI. NF?B nuclear translocation stained with labelled anti p65 antibody5 has become a widely used HCI assay. It shows good correlation to the electrophoretic mobility shift assay, but is more sensitive and requires fewer cells6. Similarly, HCI for nuclear translocation of phosphorylated ERK is superior to Western Blotting of nuclear extracts and whole cell lysates7,8. Nuclear translocation of proteins labelled by antibody staining has been successfully quantified by HCI, including ??catenin, STAT-1, STAT-3, p38 and SMAD2/39,10,7,8. TORC 1, 2 and 3 nuclear translocations were more easily measured with GFP fusion proteins11.
Antibody staining offers the advantage that the assays also run with other cell types including primary cells. But antibodies are expensive and the staining requires washing steps. Therefore, screenings are preferably based on GFP fusion proteins. Within the last few years translocation assays for a number of pharmaceutically interesting GFP fusion proteins have been developed. A selection of screening applications is commercially available from BioImage, a company that holds patents with a broad coverage of HCI translocation assays.
Another pure microscopic image based assay is the quantification of neurtite outgrowth. HCI turned out to be the ideal tool for this assay and even large scale screening has been performed as reviewed in a previous issue12. Similarly, the classical Boyden chamber cell migration assays were based on microscopic readout and have been linked to enormous workload. An important simplification of migration assays was an assay with BD fluoroblock membranes, where a plate reader quantified the fluorescence that stained cell populations transferred to the other side of the membrane. Obviously this protocol could be converted into an HCI assay. Similar to the original Boyden chamber assay, all cells were stained and the cells that reached the other side of the membrane were counted. This HCI assay was used for a number of different cell types and showed superior assay performance to the plate reader based method13,14. Moreover, the HCI method contains still greater potential, because only a single fluorescence channel is used for counting cells. Hence, multiplexing with additional fluorescent markers in the other channels is an option for additional readouts that may be used, for instance to characterise subpopulations. Other HCI migration assays have even better potential for the combination with additional endpoints. A commercially available assay is based on the phagocytosis and displacement of fluorescent beads by migrating cells, combining two fluorescence channels15. An in vitro scratch wound healing assay also using just two fluorescent channels has been used for the parallel analysis of migration and cellular morphology16.
Multiplexed endpoints
The combination of several endpoints in the same assay is a way to gain deeper and broader information from an assay. This type of multiplexing has been applied for the analysis of functional pathways, especially to distinguish between different mechanisms and to analyse connections and interdependence between pathways. As with flow cytometry, HCI allows staining in several fluorescent channels but, in addition, HCI data include parameters for localisation, texture, morphology and shape of the stained area. The use of a kinetic HCI system adds temporal patterns as an additional dimension. Independent combination of the fluorescent markers is generally possible; all endpoints show independent assay performance and need a separate validation against the respective reference assay.
A number of endpoints are accessible to address cell proliferation by HCI analysis; for a number of cell physiology parameters correlations with cell cycle phases have been analysed. Examples are the dependence of acetaminophen or cytostatic drug toxicity from cell cycle phase17,18, or cell cycle dependent expression of cyclins19. Cell cycle phase can be determined by HCI via the intensity of DNA staining analogous to FACS analysis20. In a very thorough study the cell proliferation markers cell cycle phase, BrdU incorporation, phospho-histone H3, phosphorylated retinoblastoma protein and Ki-67 antigen have been compared to plate reader based methods. The HCI endpoints showed similarly high sensitivity, but less false positive results than single endpoint ELISA based methods21.
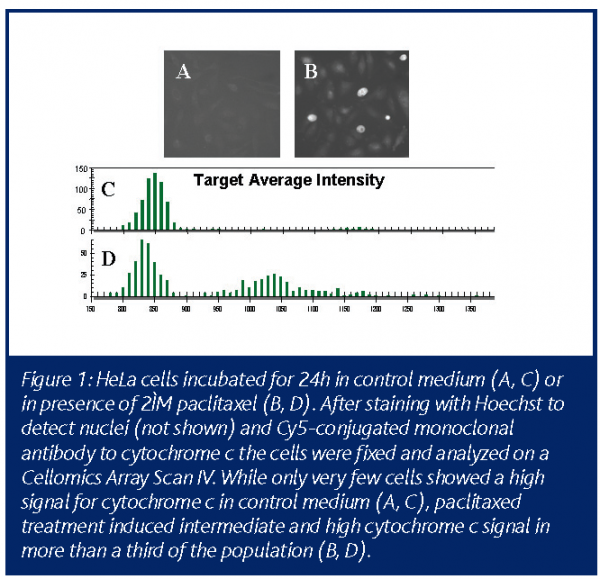
Quite a number of endpoints have been described for cell toxicity pathways and dependent on the individual project new endpoints or new combinations may be necessary. Toxicity profiles have been obtained in rat hepatocytes by kinetic analysis of multiplexed markers for nuclear morphology, cell count, membrane permeability, mitochondrial membrane potential and intracellular calcium release in the same assay22. Running several separate single endpoint assays would cost significantly more time and test substance and, in order to investigate the interdependence of mitochondrial membrane potential and short lived calcium peaks, the multiplexed kinetic HCI assay is indispensable. Other investigators performed multiplexed analysis of nuclear morphology, cell count, mitochondrial mass/potential and tubulin mass23, or of nuclear morphology, histone H3 phosphorylation, ?-tubulin structure and RSK90 phosphorylation24. The assay performance of fluorochrome-labelled inhibitors of caspases (FLICA) as indicators for caspase activity has been validated versus active caspase 3 antibody staining within the same assay, additionally versus annexin V staining, TUNEL assay, nuclear morphology and cell cycle phase25. The list of published HCI endpoints is steadily growing – an interesting new readout is the cytochrome c release assay (Figure 1). Fluorescent markers for loss of cell membrane integrity as a late toxic event are available for any fluorescence channel needed and also a number of fluorescent markers indicating certain cellular metabolic activity are accessible, especially for kinetic analysis.
Microscope based image analysis is not just a multiplexed analysis of fluorescently stained endpoints. More information is included in microscope images than the staining protocol was actually designed for. For instance, cytotoxicity markers are generally included in cell based HCI assays. The nuclear staining that is meant to identify single cells reveals cell count and nuclear morphology as indicators for cytotoxicity. Any functional assay is also multiplexed with parameters that control for artefacts such as focussing errors or fluorescent test compounds. Focussing errors of the automated microscope normally result in significantly lower nuclear staining intensity. Fluorescent test compounds enhance fluorescence intensities in parameters that should stay constant. In case of doubt it may be helpful to store the image data of an assay to analyse the images by eye.
Analysis of in vivo samples by HCI
Preclinical and clinical drug development is under pressure to obtain study data faster and to get more relevant functional information. In order to reach these goals the enhanced use of biomarkers is a current strategy and microscope based endpoints traditionally play an important role in investigating disease pathways and determining functional drug effects. HCI apparently offers the ideal combination automation and unbiased quantification by image analysis tools. As HCI analysis is possible with cytospin slides, cell smears, histology samples and even fine needle aspirates HCI analysis has already been applied in order to analyse functional in vivo responses26. The classical staining protocols usually need specific adaptations to allow optimal HCI analysis. The first HCI based pharmacodynamic (PD) assays have already been applied in clinical phase I and phase II studies to quantify PDGF receptor phosphorylation; tumour microvessel densities; vessel size and apoptosis in vessel endothelial and in tumour cells27,28,29.
The limited availability of precious biopsy material is creating an increasing need for multiplexed analysis. Also, in this case HCI appears to be the right choice and promising data have already been published. The expression and localisation pattern for 18 proteins in diseased versus healthy muscle tissue biopsies has been the first example for a highly multiplexed clinical HCI readout30.
Tissue microarrays have been generated from many types of normal and diseased tissues and are used to detect markers correlating with disease state, to understand disease mechanisms and to develop treatments. HCI systems have been applied for the functional characterisation of tissue microarrays31,32, because they provide unbiased quantification in combination with sufficient throughput.
Although HCI applications may support certain tasks in diagnostic laboratories or at pathology assessment in the future, so far no hard or software solution has shown sufficient reliability. So far the efforts necessary to control automatic analysis make the HCI approach significantly more expensive than traditional analysis.
Combination of RNA interference and HCI
Modern analysis of protein function includes protein silencing and overexpression with the help of RNA interference and cDNA expression plasmids. The functional pathways affected by interference with gene expression can be determined by HCI, which offers a number of advantages33. Aside from the option to analyse adherent cells, HCI can provide multiplexed readouts. Cotransfected fluorescent reporter genes as transfection markers allow gating on transfected cells for specific functional analysis; as a consequence functional assays are even possible in cases when transfection efficacy is low. Even more striking, protein expression levels can be quantified at the single cell level by antibody staining. The fluorescence intensity of immuno cytochemistry staining provides a good measure for protein expression on the single cell level34. This approach provides HCI analyses with great power. By multiplexing with functional endpoints a correlation between protein expression level and functional consequence is achievable on the cellular level.
For the investigation of protein function on a genome wide scale different approaches have been described and first results are expected within the coming years. Automated cell transfection protocols with cDNA, siRNA and shRNA libraries are available35,36 and HCI has been used for functional screening with high-throughput. Miniaturisation is progressing also in the field of functional assays and HCI systems are the ideal tool to screen functional effects in an ‘RNAi living-cell microarray’37.
Functional and phenotypic patterns
All approaches described above only make use of a few image parameters and endpoints. Nevertheless, any experienced scientist knows that characteristic changes in cell morphology follow treatment induced changes in cell physiology. This information can be exploited for functional assays. By HCI analysis the enormous information contained in cell morphology changes has become accessible as a pattern of hundreds of image parameters. A number of studies have been using an intelligent combination of fluorescent markers for functional and phenotypic endpoints. Highly multiplexed analysis has been successful in addressing specific functional pathways38,39, but a flexible use of the assay in other settings is limited.
In the post-genomics era with powerful biostatistical tools available there is no longer any need to exclude image analysis parameters from analysis. This way of analysing the data allows non-hypothesis driven investigation of functional assays. After genomics and proteomics, HCI is currently developing into the direction of cell-omics. First successful applications of this approach have been published. A phenotypic screen for the dose response patterns of 100 chemicals with known and unknown toxicity pathways was just based on staining DNA and actin filaments. HCI analysis delivered enormous sets of image descriptors on the single cell level and the patterns were analysed with powerful biostatistics tools. Cytological patterns were used to cluster compounds and chemicals with known similar pathways clustered together. The co-clustering with known substances was used to predict toxicity pathways for unknown compounds40. In summary, the HSC screen was not able to detect the exact molecular drug target, but data mining in image descriptors revealed the targeted functional pathways in a not-hypothesis driven analysis. Interestingly, none of the image descriptors were redundant and even descriptors without obvious biological meaning were contributing useful data41. A second research group was interested in the functional role of all known human kinases in endocytosis pathways. They transfected the HeLa cell line with an siRNA library targeting the human kinome. Both, a primary functional screen addressing viral infection via known endocytotic pathways and a secondary screen detecting the subcellular location of markers for endocytotic compartments, were run by HCI. The resulting cytological patterns were analysed with statistical tools to perform hierarchical clustering into functional clusters. Finally, detailed functional annotation was possible in all clusters that contained well characterised kinases42. So far, only specialised research groups are analysing cytological patterns. However, non-hypothesis driven functional analysis is holding a big promise. Cytological pattern analysis has not been performed within vivo samples, yet. When this type of analysis becomes feasible with reasonable throughput and costs, non-hypothesis driven functional analyses will revolutionise biosciences in a similar way that genomic analysis did recently.

References
- Conway BR, Minor LK, Xu JZ, Gunnet JW, DeBiasio R, D’Andrea MR, Rubin R, DeBiasio R, Giuliano K, DeBiasio L, Demarest KT. Quantification of G-Protein Coupled Receptor Internatilization Using G-Protein Coupled Receptor-Green Fluorescent Protein Conjugates with the ArrayScan High-Content Screening System. J. Biomol. Screen. 1999;4(2):75-86.
- Schlag BD, Lou Z, Fennell M, Dunlop J. Ligand dependency of 5-hydroxytryptamine 2C receptor internalization. J Pharmacol Exp Ther. 2004;310(3):865-70.
- Oakley RH, Hudson CC, Cruickshank RD, Meyers DM, Payne RE Jr, Rhem SM, Loomis CR. The cellular distribution of fluorescently labeled arrestins provides a robust, sensitive, and universal assay for screening G protein-coupled receptors. Assay Drug Dev Technol. 2002;1 (1 Pt 1):21-30.
- Ghosh RN, Chen YT, DeBiasio R, DeBiasio RL, Conway BR, Minor LK, Demarest KT. Cell-based, high-content screen for receptor internalization, recycling and intracellular trafficking. Biotechniques. 2000;29(1):170-5.
- Ding GJ, Fischer PA, Boltz RC, Schmidt JA, Colaianne JJ, Gough A, Rubin RA, Miller DK. Characterization and quantitation of NF-kappaB nuclear translocation induced by interleukin-1 and tumor necrosis factor-alpha. Development and use of a high capacity fluorescence cytometric system. J. Biol. Chem. 1998;273(44): 28897-905.
- Vakkila J, DeMarco RA, Lotze MT. Imaging analysis of STAT1 and NF-kappaB translocation in dendritic cells at the single cell level. J. Immunol. Methods. 2004;294 (1-2):123-34.
- Vogt A, Adachi T, Ducruet AP, Chesebrough J, Nemoto K, Carr BI, Lazo JS. Spatial analysis of key signaling proteins by high-content solid-phase cytometry in Hep3B cells treated with an inhibitor of Cdc25 dual-specificity phosphatases. J. Biol. Chem. 2001;276(23):20544-50.
- Partridge EA, Le Roy C, Di Guglielmo GM, Pawling J, Cheung P, Granovsky M, Nabi IR, Wrana JL, Dennis JW. Regulation of cytokine receptors by Golgi N-glycan processing and endocytosis. Science. 2004;306(5693):120-4.
- Borchert KM, Galvin RJ, Frolik CA, Hale LV, Halladay DL, Gonyier RJ, Trask OJ, Nickischer DR, Houck KA. High-content screening assay for activators of the Wnt/Fzd pathway in primary human cells. Assay Drug Dev Technol. 2005;3(2):133-41.
- Parrish-Novak J, Xu W, Brender T, Yao L, Jones C, West J, Brandt C, Jelinek L, Madden K, McKernan PA, Foster DC, Jaspers S, Chandrasekher YA. Interleukins 19, 20, and 24 signal through two distinct receptor complexes. Differences in receptor-ligand interactions mediate unique biological functions. J. Biol. Chem. 2002;277(49): 47517-23.
- Bittinger MA, McWhinnie E, Meltzer J, Iourgenko V, Latario B, Liu X, Chen CH, Song C, Garza D, Labow M. Activation of cAMP response element-mediated gene expression by regulated nuclear transport of TORC proteins. Curr. Biol. 2004;14(23):2156-61.
- Simpson PB. Getting a handle on neuronal behaviour in culture. Eur. Pharm. Rev. 2005; 2: 56-64.
- Mastyugin V, McWhinnie E, Labow M, Buxton F. A quantitative high-throughput endothelial cell migration assay. J. Biomol. Screen. 2004;9(8):712-8.
- Richards GR, Millard RM, Leveridge M, Kerby J, Simpson PB. Quantitative assays of chemotaxis and chemokinesis for human neural cells. Assay Drug Dev. Technol. 2004;2(5):465-72.
- Nam JS, Ino Y, Kanai Y, Sakamoto M, Hirohashi S. 5-aza-2′-deoxycytidine restores the E-cadherin system in E-cadherin-silenced cancer cells and reduces cancer metastasis. Clin Exp Metastasis. 2004;21(1):49-56.
- Yarrow JC, Feng Y, Perlman ZE, Kirchhausen T, Mitchison TJ. Phenotypic screening of small molecule libraries by high throughput cell imaging. Comb Chem High Throughput Screen. 2003 Jun;6(4):279-86.
- Cai Q, Dmitrieva NI, Michea LF, Rocha G, Ferguson D, Burg MB. Toxicity of acetaminophen, salicylic acid, and caffeine for first-passage rat renal inner medullary collecting duct cells. J Pharmacol Exp Ther. 2003;306(1):35-42.
- Pozarowski P, Huang X, Gong RW, Priebe W, Darzynkiewicz Z. Simple, semiautomatic assay of cytostatic and cytotoxic effects of antitumor drugs by laser scanning cytometry: effects of the bis-intercalator WP631 on growth and cell cycle of T-24 cells. Cytometry A. 2004;57(2):113-9.
- Takita M, Furuya T, Sugita T, Kawauchi S, Oga A, Hirano T, Tsunoda S, Sasaki K. An analysis of changes in the expression of cyclins A and B1 by the cell array system during the cell cycle: comparison between cell synchronization methods. Cytometry A. 2003;55(1):24-9.
- Kamentsky LA, Burger DE, Gershman RJ, Kamentsky LD, Luther E. Slide-based laser scanning cytometry. Acta Cytol. 1997;41(1):123-43.
- Gasparri F, Mariani M, Sola F, Galvani A. Quantification of the proliferation index of human dermal fibroblast cultures with the ArrayScan high-content screening reader. J Biomol Screen. 2004;9(3):232-43.
- Haskins JR, Rowse P, Rahbari R, de la Iglesia FA. Thiazolidinedione toxicity to isolated hepatocytes revealed by coherent multiprobe fluorescence microscopy and correlated with multiparameter flow cytometry of peripheral leukocytes. Arch Toxicol. 2001;75(7):425-38.
- Vogt A, Kalb EN, Lazo JS. A scalable high-content cytotoxicity assay insensitive to changes in mitochondrial metabolic activity. Oncol Res. 2004;14(6):305-14.
- Minguez JM, Giuliano KA, Balachandran R, Madiraju C, Curran DP, Day BW. Synthesis and high content cell-based profiling of simplified analogues of the microtubule stabilizer (+)-discodermolide. Mol Cancer Ther. 2002;1(14):1305-13.
- Pozarowski P, Huang X, Halicka DH, Lee B, Johnson G, Darzynkiewicz Z. Interactions of fluorochrome-labeled caspase inhibitors with apoptotic cells: a caution in data interpretation. Cytometry A. 2003;55(1):50-60.
- Mocellin S, Wang E, Panelli M, Rossi CR, Marincola FM. Use of laser scanning cytometry to study tumor microenvironment. Histol Histopathol. 2003;18(2): 609-15.
- Davis DW, Shen Y, Mullani NA, Wen S, Herbst RS, O’Reilly M, Abbruzzese JL, McConkey DJ. Quantitative analysis of biomarkers defines an optimal biological dose for recombinant human endostatin in primary human tumors. Clin Cancer Res. 2004;10(1 Pt 1):33-42.
- Heymach JV, Desai J, Manola J, Davis DW, McConkey DJ, Harmon D, Ryan DP, Goss G, Quigley T, Van den Abbeele AD, Silverman SG, Connors S, Folkman J, Fletcher CD, Demetri GD. Phase II study of the antiangiogenic agent SU5416 in patients with advanced soft tissue sarcomas. Clin Cancer Res. 2004;10(17):5732-40.
- Davis DW, Takamori R, Raut CP, Xiong HQ, Herbst RS, Stadler WM, Heymach JV, Demetri GD, Rashid A, Shen Y, Wen S, Abbruzzese JL, McConkey DJ. Pharmacodynamic analysis of target inhibition and endothelial cell death in tumors treated with the vascular endothelial growth factor receptor antagonists SU5416 or SU6668. Clin Cancer Res. 2005;11(2 Pt 1):678-89.
- Schubert W. Topological proteomics, toponomics, MELK-technology. Adv Biochem Eng Biotechnol. 2003;83:189-209.
- Balmer D, Goldstine J, Rao YM, LaSalle JM. Elevated methyl-CpG-binding protein 2 expression is acquired during postnatal human brain development and is correlated with alternative polyadenylation. J Mol Med. 2003;81(1):61-8.
- Samaco RC, Nagarajan RP, Braunschweig D, LaSalle JM. Multiple pathways regulate MeCP2 expression in normal brain development and exhibit defects in autism-spectrum disorders. Hum Mol Genet. 2004;13(6):629-39.
- Bhawe KM, Blake RA, Clary DO, Flanagan PM. An automated image capture and quantitation approach to identify proteins affecting tumor cell proliferation. J Biomol Screen. 2004;9(3):216-22.
- Erfle H, Simpson JC, Bastiaens PI, Pepperkok R. siRNA cell arrays for high-content screening microscopy. Biotechniques. 2004;37(3):454-8, 460, 462.
- Ziauddin J, Sabatini DM. Microarrays of cells expressing defined cDNAs. Nature. 2001;411(6833):107-10.
- Silva JM, Mizuno H, Brady A, Lucito R, Hannon GJ. RNA interference microarrays: high-throughput loss-of-function genetics in mammalian cells. Proc Natl Acad Sci U S A. 2004;101(17):6548-52
- Wheeler DB, Bailey SN, Guertin DA, Carpenter AE, Higgins CO, Sabatini DM. RNAi living-cell microarrays for loss-of-function screens in Drosophila melanogaster cells. Nat Methods. 2004;1(2):127-32.
- Walmod PS, Berezin A, Gallagher HC, Gravemann U, Lepekhin EA, Belman V, Bacon CL, Nau H, Regan CM, Berezin V, Bock E. Automated in vitro screening of teratogens. Toxicol Appl Pharmacol. 2002;181(1):1-15.
- Giuliano KA. High-content profiling of drug-drug interactions: cellular targets involved in the modulation of microtubule drug action by the antifungal ketoconazole. J Biomol Screen. 2003;8(2):125-35.
- Perlman ZE, Slack MD, Feng Y, Mitchison TJ, Wu LF, Altschuler SJ. Multidimensional drug profiling by automated microscopy. Science. 2004;306(5699): 1194-8.
- Mitchison TJ. Small-molecule screening and profiling by using automated microscopy. Chembiochem. 2005;6(1):33-9.
- Pelkmans L, Fava E, Grabner H, Hannus M, Habermann B, Krausz E, Zerial M. Genome-wide analysis of human kinases in clathrin- and caveolae/raft-mediated endocytosis. Nature. 2005;436(7047):78-86.




