Isothermal titration calorimetry and thermal shift assay in drug design
Posted: 20 June 2011 |
Isothermal titration calorimetry (ITC) is a method of choice in the pharmaceutical industry for determination of equilibrium binding enthalpy, entropy, and the Gibbs free energy. The method is very powerful for determination of intrinsic binding parameters that could be used in structure-energetics correlations. Here we discuss how to overcome several limitations of ITC. First, it is easy to complement ITC results with thermal shift assay (TSA) in order to avoid the narrow window of ITC Kd measurements. Second, several examples are provided on how to determine intrinsic enthalpy of binding and Kd. Third, ITC and TSA Kds are compared with enzymatic inhibition methods.
Isothermal titration calorimetry is a wellestablished method for determining the association constant and other thermodynamic parameters of intermolecular interactions in aqueous solutions. The technique has been widely used to study interactions between very different molecular species such as proteins, DNA, RNA, lipids, drug leads, metal ions and many other chemical substances as previously reviewed1,2.
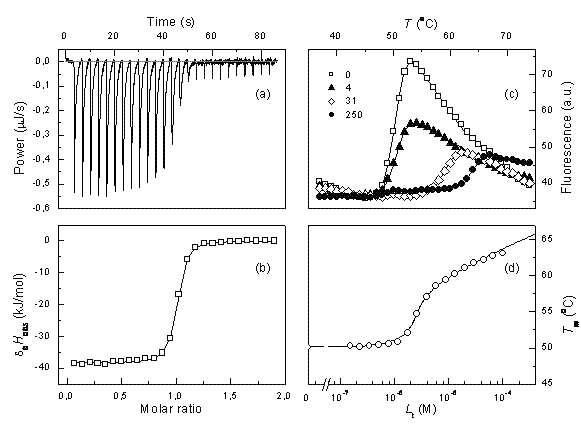

Figure 1 Comparison of ITC and TSA data for lead compound 3b binding to the N-terminal domain of Hsp90 target protein4. Left panels show ITC data, right panels – TSA data. Upper panels (a) and (c) show raw data while lower panels show the dosing curves
Isothermal titration calorimetry (ITC) is a method of choice in the pharmaceutical industry for determination of equilibrium binding enthalpy, entropy, and the Gibbs free energy. The method is very powerful for determination of intrinsic binding parameters that could be used in structure-energetics correlations. Here we discuss how to overcome several limitations of ITC. First, it is easy to complement ITC results with thermal shift assay (TSA) in order to avoid the narrow window of ITC Kd measurements. Second, several examples are provided on how to determine intrinsic enthalpy of binding and Kd. Third, ITC and TSA Kds are compared with enzymatic inhibition methods.
Isothermal titration calorimetry is a wellestablished method for determining the association constant and other thermodynamic parameters of intermolecular interactions in aqueous solutions. The technique has been widely used to study interactions between very different molecular species such as proteins, DNA, RNA, lipids, drug leads, metal ions and many other chemical substances as previously reviewed1,2.
The early stages of high-throughput drug discovery in the pharmaceutical industry are usually based on selecting the highest affinity compounds that bind to and inhibit the target protein function. However, the affinity or association constant (Kb = 1/Kd) alone provides limited information on the forces driving the binding event. Chemical thermodynamics teaches us that the equilibrium binding (association) constant Kb is related to the Gibbs free energy of binding:
![]()
![]()
The Gibbs free energy is made from enthalpic and entropic contributions to the reaction. From a single ITC experiment one can obtain all three thermodynamic parameters including Gibbs free energy (ΔbG), enthalpy (ΔbH) and entropy (ΔbS).
![]()
![]()
Compounds with similar affinities often have very different enthalpic and entropic contributions due to the enthalpy-entropy compensation phenomenon. Such compounds have different reasons for binding. In the stages of optimising the lead compound, it is important to take into consideration as many energetic contributions to the binding reaction as possible. But the enthalpic term is the most important. It is a direct measure of the net change in the number and strength of the non-covalent bonds on going from the free to the bound state. ITC provides a very good tool to directly measure the enthalpy of interaction. However, one has to be careful with the interpretation of the enthalpic data. It is important to carry out all necessary controls and dissect the intrinsic enthalpy from other contributions occurring in solution due to linked reactions. Most commonly, there are large enthalpic contributions from linked protonation reactions that depend on buffer and pH. Both the protein and ligand may be protonated or deprotonated upon binding. The methodology of dissection has been well described3 but the interpretation of various binding reactions described in the literature is still often incomplete. The structure-thermodynamics correlations cannot be applied without detailed dissection of such contributing reactions.
Here we will discuss several issues/aspects of ITC that need to be understood and checked in every application.
Narrow window of Kb
ITC data can be reliably fit and the Kb determined only if the multiplication product between the observed Kb and the molar concentration of the protein is between about 5 and 500. If the usual protein concentration in the experiments is 10-5 M, then one can determine the Kbs in the range of 5×105 to 5×107M-1. The affinities of lead compounds often fall out of this range. In such cases, we argue that the thermal shift assay (TSA, also called ThermoFluor or differential scanning fluorimetry) is a method of choice to confirm or correct ITC data. Figure 1 shows a comparison of typical ITC and TSA data.


Figure 1 Comparison of ITC and TSA data for lead compound 3b binding to the N-terminal domain of Hsp90 target protein4. Left panels show ITC data, right panels – TSA data. Upper panels (a) and (c) show raw data while lower panels show the dosing curves
Determination of the intrinsic enthalpy
In order to draw any enthalpic energeticsstructure correlations, the determined binding enthalpy must be intrinsic. Notable is that any linked protonation event in tris buffer could change the enthalpy by about 48 kJ/mol (heat of deprotonation of tris buffer, ΔbHbuffer). This value is comparable to the enthalpies of binding of most ligands that are in the range of 0-100 kJ/mol.
The intrinsic enthalpy is related to the observed enthalpy:
![]()
![]()
where n is the number of linked protonation events and ΔbHcomplex is the enthalpy of protein and/or ligand binding-linked protonation.
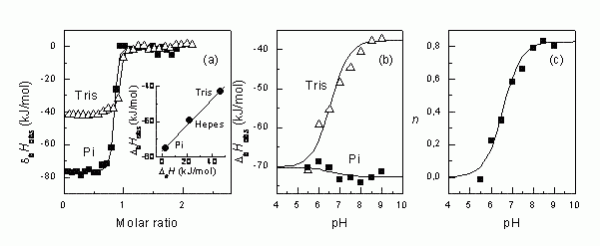

Figure 2The steps that are necessary to determine the intrinsic binding enthalpy. First, the binding ITC experiment must be conducted in several buffers (a). Second, if there is difference in observed enthalpies, then it is desirable to repeat ITC experiments in several buffers at multiple pHs (b). Third, the numbers of protons transferred are plotted as a function of pH, determining the pKa of the linked protonation reaction8
Figure 2 shows how different the observed enthalpies of binding may be indifferent buffers. In tris buffer, the ΔbHobs is about -41 kJ/mol and in phosphate buffer, it is -77 kJ/mol at pH 8.0. The difference is due to the linked protonation events as explained in Figure 3.
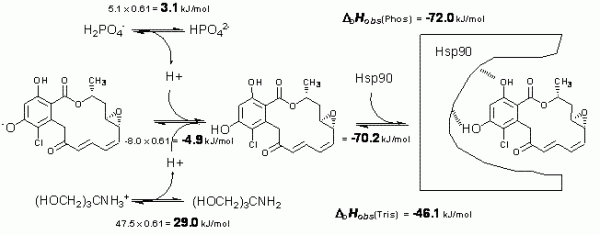

Figure 3 Schematic depiction of linked protonation events occurring in aqueous buffer solution upon radicicol binding to Hsp90 at pH 8.0. Top left numbers evaluate enthalpic contribution of n = 0.61 proton deprotonation from phosphate. Top right number shows the observed enthalpy in phosphate. Middle left number is the determined enthalpy of radicicol protonation. The right middle number is the determined intrinsic binding enthalpy. Lower numbers are the observed enthalpies in tris buffer
Figure 3 shows enthalpic contributions to the observed enthalpy of radicicol binding to Hsp90 in phosphate and tris buffers. After the application of additivity principles, we estimate that the intrinsic enthalpy of binding at 25°C is equal to -72 kJ/mol. This is the enthalpy of greatest interest and only this value may be used in structureenergetics correlations.
Determination of the intrinsic dissociation constant
Similarly to the observed enthalpy, the observed dissociation constant will also depend on pH if there are linked protonation events. This observed Kd must be determined at multiple pHs. Quite often there are no conditions where the intrinsic Kd may be observed experimentally directly and match the observed Kd. ITC is especially useful because it can determine intrinsic Kd in the same series of experiments as used to determine the intrinsic binding enthalpies.
If ligand binding is linked to the binding of a single proton, then the observed binding con – stant (Kobs) and the intrinsic binding constant to the protonated protein form (Kintr) are related by:
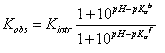
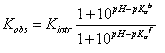
where Ka b and Ka f are the proton dissociation constants from the liganded and unliganded protein, respectively.
Figure 4 shows pH dependencies of observed Kd for two inhibitor-target protein systems, namely, ethoxzolamide – carbonic anhydrase (CA II) and radicicol – Hsp90. At pH 7.0, the difference between the intrinsic and observed Kd is about 20× for CA II. However, at other pHs the difference is even greater and the intrinsic Kd never matches the observed Kd and cannot be observed experimentally at any pH. The U-shape is caused by two linked protonation events: protonation of the enzyme and deprotonation of the inhibitor. The Hsp90 – radicicol system has only one linked protonation event, most likely of the radicicol as explained in Figure 3. Therefore, at low pH the two Kds match each other, while at alkaline pH the observed Kd is continuously weaker.
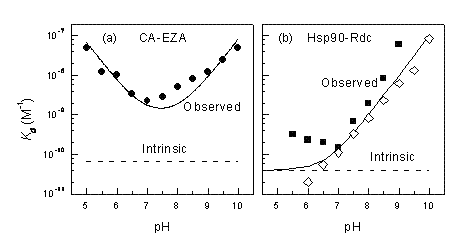

Figure 4 Observed and intrinsic Kds are shown as a function of pH for carbonic anhydrase – ethoxzolamide (a) and Hsp90 – radicicol systems. Panel (b) compares experimental datapoints obtained by ITC (filled squares) and TSA (open rhombs). Below pH 7 the ITC reached its maximum Kd determination capability. TSA measurements at low pH are more accurate than ITC8-10
Do the ITC and TSA Kds match the inhibition constant of the enzymatic activity Ki?
In high throughput screening, pharmaceutical companies usually employ methods that directly observe enzyme inhibition. Supposedly, such methods could provide more accurate estimation of inhibition efficiency than the biophysical binding measurements such as ITC and TSA. Figure 5 compares the Kds obtained by ITC and TSA with direct inhibition measurements7.
Generally, there is quite good match between the Kds and Kis determined in two different laboratories on different protein preparations and employing very different approaches. The most significant mismatch reached up to about 10 fold. In thermodynamic terms, such difference is quite insignificant and could probably be attributed to random error of all three methods. Therefore, both ITC and TSA are suitable for screening ligands.
Discussion and conclusions
ITC is a powerful technique to determine the thermodynamic parameters of binding. However, sometimes despite very nice-looking data, authors only obtain observed thermodynamic parameters11. Such functional group additivity schemes are not fully described because the authors only use the observed thermodynamic parameters. Even if the experiments are carried in the same buffer at same pH, the compounds may have ionisable groups with different pKas as is in the case with sulfonamide inhibitors. Therefore, in order to draw true energetics – structure correlations, one must carry out many ITC experiments. However, for the first approximation as done by Scott et al11, compound observed enthalpies and observed Kds may be ranked by a single ITC experiment.
References
- Ladbury JE (2010) Calorimetry as a tool for understanding biomolecular interactions and an aid to drug design. Biochem Soc Trans 38:888–893
- Velazquez Campoy A, Freire E (2005) Itc in the postgenomic era…? priceless. Biophys Chem 115:115–24
- Baker BM, Murphy KP (1996) Evaluation of linked protonation effects in protein binding reactions using isothermal titration calorimetry. Biophysical Journal 71(4):2049–55
- Cikotiene I, et al. (2009) 5-aryl-4-(5-substituted-2,4- dihydroxyphenyl)-1,2,3-thiadiazoles as inhibitors of hsp90 chaperone. Bioorg Med Chem Lett 19:1089–1092
- Matulis D, Kranz JK, Salemme FR, Todd MJ (2005) Thermodynamic stability of carbonic anhydrase: measurements of binding affinity and stoichiometry using thermofluor. Biochemistry 44:5258–66
- Cimmperman P, et al. (2008) A quantitative model of thermal stabilization and destabilization of proteins by ligands. Biophys J 95:3222–3231
- Baranauskiene L, et al. (2010) Inhibition and binding studies of carbonic anhydrase isozymes i, ii and ix with benzimidazo[1,2-c][1,2,3]thiadiazole-7- sulphonamides. J Enzyme Inhib Med Chem 25:863–870
- Zubriene A, et al. (2010) Thermodynamics of radicicol binding to human hsp90 alpha and beta isoforms. Biophys Chem 152:153–163
- Krishnamurthy VM, et al. (2008) Carbonic anhydrase as a model for biophysical and physical-organic studies of proteins and protein-ligand binding. Chem Rev 108:946–1051
- Matulis D, Todd MJ (2004) Thermodynamics – Structure Correlations of Sulfonamide Inhibitor Binding to Carbonic Anhydrase, eds Ladbury JE, Doyle ML (Wiley), pp 107–132
- Scott AD, et al. (2009) Thermodynamic optimisation in drug discovery: a case study using carbonic anhydrase inhibitors. ChemMedChem 4:1985–9




