HOX genes: HOX transcription factors as biomarkers in cancer
Posted: 19 October 2011 |
The HOX genes are a family of closely related transcription factors that help to define the identity of cells and tissues during embryonic development and which are also frequently deregulated in cancer, where they have been shown to promote cell survival and proliferation. The high level of cancer-associated HOX expression and the pro-oncogenic functions of these genes make them strong candidates for biomarkers in multiple roles including diagnosis, prognosis, drug sensitivity and drug resistance. The HOX genes are a family of homeodomaincontaining transcription factors that were first identified as determinates of cell and tissue identity in early development, although they are now also known to function in adult stem cell renewal and differentiation. A series of duplication events is thought to have given rise to the four separate clusters of HOX genes found in vertebrates, with each cluster consisting of a group of closely linked members that often share enhancer regions. These clusters are named A, B, C and D, and together they contain the 39 HOX genes found in mammals. Each gene within a cluster is labelled with a number according to their relative position in the chromosome, so for example HOXB1 is the 3’ most member of the B cluster, and HOXB13 is the 5’ most member. The linkage of genes within each cluster is closely reflected in both their temporal and spatial order of expression in the embryo, with the 3’ genes being expressed more anteriorly and earlier than their 5’ neighbours. The relative position within the cluster is also reflected in the co-factor interactions, DNA binding specificity and regulation of each member.
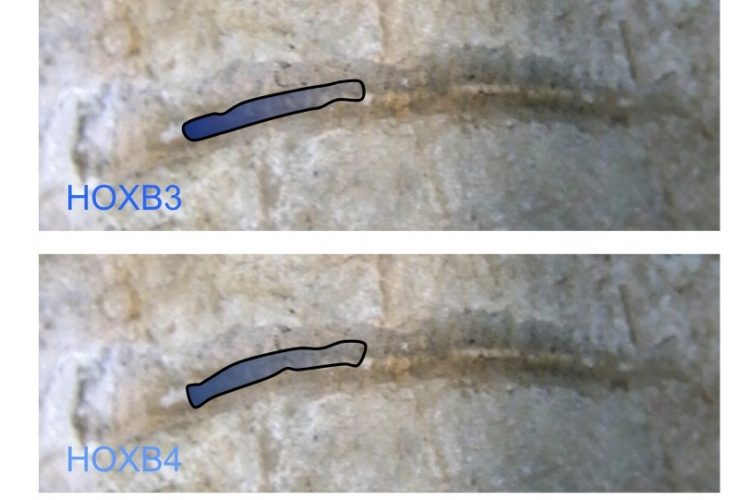

FIGURE 1 Nested HOX gene expression along the anterior to posterior axis. The expression domains of members of the HOXB group are illustrated, superimposed on the spinal cord of an early vertebrate. The combined expression of HOX genes in defined spatial positions is a key determinate of cell and tissue identity
The HOX genes are a family of closely related transcription factors that help to define the identity of cells and tissues during embryonic development and which are also frequently deregulated in cancer, where they have been shown to promote cell survival and proliferation. The high level of cancer-associated HOX expression and the pro-oncogenic functions of these genes make them strong candidates for biomarkers in multiple roles including diagnosis, prognosis, drug sensitivity and drug resistance.
The HOX genes are a family of homeodomaincontaining transcription factors that were first identified as determinates of cell and tissue identity in early development, although they are now also known to function in adult stem cell renewal and differentiation1. A series of duplication events is thought to have given rise to the four separate clusters of HOX genes found in vertebrates, with each cluster consisting of a group of closely linked members that often share enhancer regions. These clusters are named A, B, C and D, and together they contain the 39 HOX genes found in mammals2. Each gene within a cluster is labelled with a number according to their relative position in the chromosome, so for example HOXB1 is the 3’ most member of the B cluster, and HOXB13 is the 5’ most member3. The linkage of genes within each cluster is closely reflected in both their temporal and spatial order of expression in the embryo, with the 3’ genes being expressed more anteriorly and earlier than their 5’ neighbours (Figure 1). The relative position within the cluster is also reflected in the co-factor interactions, DNA binding specificity and regulation of each member2.
HOX genes also deregulated in a number of cancers
In addition to a role in development, and subsequently in stem cell differentiation, the HOX genes are also frequently deregulated in a number of cancers including melanoma, mesothelioma, and lung, kidney, prostate, ovarian and breast cancer4. Their function in oncogenesis is still unclear; however, it is apparent that the great complexity of HOX function in development is also reflected in oncogenesis, with some HOX genes functioning as tumour suppressors and others as oncogenes. The best known examples of both have been identified in breast cancer, where HOXA5 is known to function as a tumour suppressor5, at least in part through activating the transcription of the key tumour suppressor gene TP536. Conversely, members of the closely related HOXB genes including HOXB5 and HOXB7 are oncogenic through mechanisms which include an up regulation of FGF27, and the promotion of epithelial to mesenchymal transition8. Exactly how such similar transcription factors can have opposing functions is also unclear, although it may be related to differential co-factor binding and consequently differential regulation of target genes. Known co-factors include members of the PBX, MEIS and PREP families of transcription factors, all of which can influence the binding selectivity of HOX proteins and their action as either a suppressor or activator of transcription1. As an additional complexity, HOX proteins can also regulate transcription through binding to DNA as monomers9.
Whilst different HOX genes can have individual, specific functions in embryonic development, there is generally a high level of functional redundancy, especially with regards to fundamental and highly conserved patterning events such as anterior-posterior pattering of the spine and hindbrain10,11. This is also true in cancer, where a similar oncogenic function is common to a number of HOX genes, especially HOXB1 through to HOXB911,12. The highly complex expression of HOX genes in different cancers gives rise to the possibility of a ‘fingerprint’ that can be used to distinguish different types of cancer, and also to potentially diagnose cancer based on the presence of specific HOX proteins in cells or in bodily fluids. Furthermore, the diverse range of functions that HOX genes have in cancer also raises the possibility of using them as prognostic markers and / or predictive markers for the response to treatment. As yet this is an under-explored area of research, but here I have reviewed some of the key findings to date that illustrate the potential of HOX genes as biomarkers in cancer.
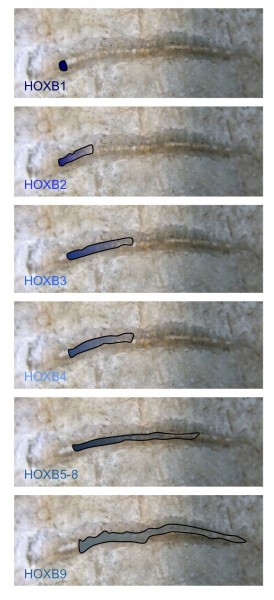

FIGURE 1 Nested HOX gene expression along the anterior to posterior axis. The expression domains of members of the HOXB group are illustrated, superimposed on the spinal cord of an early vertebrate. The combined expression of HOX genes in defined spatial positions is a key determinate of cell and tissue identity
Leukaemia
HOX genes are well established as having an oncogenic role in a number of different leukemias, and their expression is frequently associated with a poor prognosis13. One of the most striking examples is HOXA9, the up regulation of which is associated with a poor outcome in acute myeloid leukaemia (AML), and when combined with the expression of a number of other HOX genes is able to predict the occurrence of an unfavourable cytology with 95 per cent specificity14. HOXA9 is required for the survival of malignant cells15, and is also frequently involved in the formation of HOX fusion genes that arise through chromosomal rearrangement and which are key drivers of many haematological malignancies13. It is also interesting to note that the expression of HOXA9 gradually increases in normal adult blood cells with age, although the functional significance of this is as yet unknown16.
Breast cancer
Our work, as that of others, has shown that HOX genes are frequently expressed in breast tumours at a far higher level than in normal breast tissue (Figure 2). Evidence for a functional role of this over expression has been provided in part by studies showing HOXB7 to have a number of key roles in breast cancer, impacting both on overall survival and drug resistance. The expression of HOXB7 by breast cancer cells is associated with enhanced epithelial to mesenchymal transition, a key event in metastasis17. HOXB7 is correspondingly associated with poor survival (although only in Her2 positive patients)17. More recent work has shown that HOXB7 is also a potential predictor of tamoxifen resistance in ERα positive breast cancer; HOXB7 can mediate tamoxifen resistance by up regulating EGFR through direct binding to the gene’s promoter18.
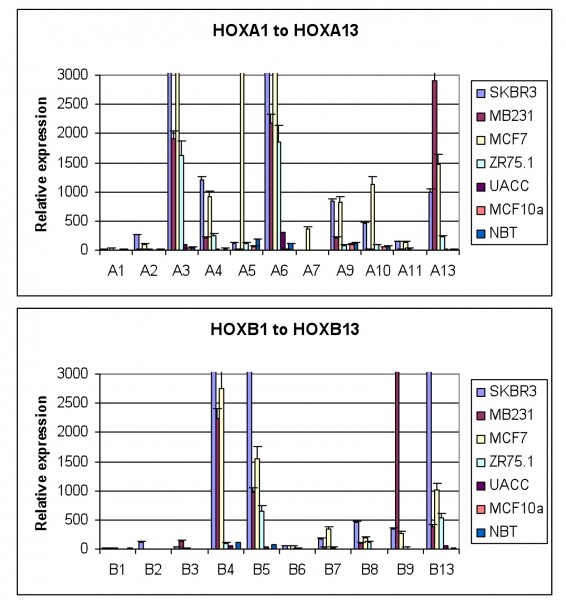

FIGURE 2 HOX gene expression in breast cancer derived cell lines and in normal breast tissue. The expression of each gene was determined by semi-quantitative PCR and is shown relative to the house keeping gene GAPDH (x10000). The values shown are the mean of three independent experiments and the error bars represent the SEM. NBT – normal breast tissue. Everyone of the breast tumour derived cell lines show significant up regulation of many HOX genes whilst there is very little HOX expression in normal breast tissue, or in MCF10a, a cell line derived from non-malignant mammary cells
The multiple functions of HOX genes in cancer cell survival and proliferation make them potential therapeutic targets, and HOX expression is a potential marker for sensitivity to these therapies. However, the therapeutic targeting of HOX proteins is complicated by opposing functionality and functional redundancy, combined with a lack of ligand binding sites. One possible solution is to target multiple groups of HOX genes in a way that also singles out specific HOX functions, something that could potentially be achieved by disrupting the binding of HOX proteins to specific cofactors. To date this has only been possible for the PBX co-factor that can bind to HOX proteins numbered 1 to 91219-22. PBX increases the nuclear translocation of HOX proteins and also influences the selection of DNA binding sites23,24. Its interaction with HOX is mediated by a highly conserved hexapeptide region on HOX proteins23-26 and previous studies have shown that a synthetic peptide consisting of these amino acids and a short polyarginine sequence, known as HXR9, is capable of blocking the interaction between HOX and PBX proteins both in vitro and in vivo. HXR9 causes apoptosis in a number of cancers including melanoma12, myeloma21, and kidney19, non-small cell lung20, and ovarian cancer22. We have found that HXR9 also causes apoptosis in cell lines derived from different breast cancers, and that there is a strong correlation between the average expression of HOX genes HOXB1 through HOXB9 and the sensitivity of cell killing by HXR9. Hence the expression of specific groups of HOX genes may predict which cancers will respond to HXR9, or its derivatives, or indeed other treatment modalities that target HOX function.
Ovarian cancer
A role for HOXB7 is also implied in ovarian cancer by the presence of circulating autologous antibodies to its protein product, a finding that also suggests a possible role for this immune response as a diagnostic tool27,28. Interestingly, autoantibodies to the closely related HOXA7 protein are a potential biomarker of this disease29, and analysis of HOX gene expression in tumours has indicated that HOXA10 is a promising prognostic marker for ovarian cancer as its over expression is strongly correlated with poor survival30. The analysis of HOX expression also extends to the genetic level in ovarian cancer as promoter methylation is often important in the global regulation of HOX clusters. In particular, methylation of the HOXA11 promoter is strongly associated with poor outcome in ovarian cancer, suggesting a possible role for DNA methylation as a prognostic marker in ovarian cancer31.
Engrailed-2 (EN2)
Engrailed-2 (EN2) is very closely related to the HOX transcription factors, being characterised by a homeodomain and having similar co-factor binding activities, but it is not contained within the four HOX clusters described above32. Like the HOX genes, EN2 also shows a very high degree of functional conservation during development, being required for patterning of the brain33 and limbs32. However, unlike many of the HOX genes, EN2 is not expressed in adult cells (with the possible exception of the purkinje neurons34) and but it is re-expressed in both breast35 and prostate36 cancer. In the former, EN2 has been shown to function as an oncogene, as forcing its expression in non-malignant mammary cells induces a malignant phenotype including increased cell proliferation and a loss of contact dependence35. The absence of EN2 expression in normal adult cells together with its oncogenic function makes it an ideal candidate for a cancer biomarker. Indeed, our work has shown that EN2 protein is secreted from prostate tumours and can be detected in urine36. This study compared EN2 protein concentrations in the urine of prostate cancer patients and men with non-cancerous conditions of the prostate such as benign hypertrophy, and revealed significantly increased EN2 levels in cancer. These results suggest that EN2 is a potential diagnostic marker for prostate cancer with a sensitivity and specificity of 66 per cent and 82 per cent, respectively.
Conclusions
The relatively low expression of HOX genes in normal adult cells combined with their frequent up regulation and pro-survival function in cancer cells make them strong candidates for biomarkers. This potential is enhanced further by the complexity of their expression patterns which varies from one cancer to another, and between sub-types. Some, if not all, of this variation reflects specific functions of HOX genes and thus there is enormous potential for the HOX genes and their products to act as biomarkers in a wide range of contexts. In this review, I have summarised some of the existing uses and potential uses of HOX genes as biomarkers, but this is likely to be only the tip of a very useful resource.
References
- Moens, C.B. and L. Selleri, Hox cofactors in vertebrate development. Dev Biol, 2006. 291(2): p. 193-206
- Hoegg, S. and A. Meyer, Hox clusters as models for vertebrate genome evolution. Trends Genet, 2005. 21(8): p. 421-4
- Scott, M.P., A rational nomenclature for vertebrate homeobox (HOX) genes. Nucleic Acids Res, 1993. 21(8): p. 1687-8 4. Shah, N. and S. Sukumar, The Hox genes and their roles in oncogenesis. Nat Rev Cancer, 2010. 10(5): p. 361-71
- Chen, H., et al., HOXA5 acts directly downstream of retinoic acid receptor beta and contributes to retinoic acid-induced apoptosis and growth inhibition. Cancer Res, 2007. 67(17): p. 8007-13
- Raman, V., et al., Compromised HOXA5 function can limit p53 expression in human breast tumours. Nature, 2000. 405(6789): p. 974-8
- Care, A., et al., Transduction of the SkBr3 breast carcinoma cell line with the HOXB7 gene induces bFGF expression, increases cell proliferation and reduces growth factor dependence. Oncogene, 1998. 16(25): p. 3285-9
- Wu, X., et al., HOXB7, a homeodomain protein, is overexpressed in breast cancer and confers epithelialmesenchymal transition. Cancer Res, 2006. 66(19): p. 9527-34
- Galant, R., C.M. Walsh, and S.B. Carroll, Hox repression of a target gene: extradenticle-independent, additive action through multiple monomer binding sites. Development, 2002. 129(13): p. 3115-26
- Di-Poi, N., et al., Additive and global functions of HoxA cluster genes in mesoderm derivatives. Dev Biol, 2010. 341(2): p. 488-98
- Lappin, T.R., et al., HOX genes: seductive science, mysterious mechanisms. Ulster Med J, 2006. 75(1): p. 23-31
- Morgan, R., et al., Antagonism of HOX/PBX dimer formation blocks the in vivo proliferation of melanoma. Cancer Res, 2007. 67(12): p. 5806-13
- McGonigle, G.J., T.R. Lappin, and A. Thompson, Grappling with the HOX network in hematopoiesis and leukemia. Front Biosci, 2008. 13: p. 4297-308
- Andreeff, M., et al., HOX expression patterns identify a common signature for favorable AML. Leukemia, 2008. 22(11): p. 2041-7
- Faber, J., et al., HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood, 2009. 113(11): p. 2375-85
- Morgan, R., et al., HOXA9 expression increases with age in human haemopoietic cells. Leuk Res, 2005. 29(10): p. 1221-2
- Chen, H., et al., Hoxb7 inhibits transgenic HER-2/ neu-induced mouse mammary tumor onset but promotes progression and lung metastasis. Cancer Res, 2008. 68(10): p. 3637-44
- Jin, K., et al., Breast Cancer Special Feature: The HOXB7 protein renders breast cancer cells resistant to tamoxifen through activation of the EGFR pathway. Proc Natl Acad Sci U S A, 2011
- Shears, L., et al., Disrupting the interaction between HOX and PBX causes necrotic and apoptotic cell death in the renal cancer lines CaKi-2 and 769-P. J Urol, 2008. 180(5): p. 2196-201
- Plowright, L., et al., HOX transcription factors are potential therapeutic targets in non-small-cell lung cancer (targeting HOX genes in lung cancer). Br J Cancer, 2009. 100(3): p. 470-5
- Daniels, T.R., et al., Disruption of HOX activity leads to cell death that can be enhanced by the interference of iron uptake in malignant B cells. Leukemia, 2010. 24(9): p. 1555-65
- Morgan, R., et al., Targeting HOX and PBX transcription factors in ovarian cancer. BMC Cancer, 2010. 10: p. 89
- Phelan, M.L., R. Sadoul, and M.S. Featherstone, Functional differences between HOX proteins conferred by two residues in the homeodomain Nterminal arm. Mol Cell Biol, 1994. 14(8): p. 5066-75
- Piper, D.E., et al., Structure of a HoxB1-Pbx1 heterodimer bound to DNA: role of the hexapeptide and a fourth homeodomain helix in complex formation. Cell, 1999. 96(4): p. 587-97
- Knoepfler, P.S., et al., Meis1 and pKnox1 bind DNA cooperatively with Pbx1 utilizing an interaction surface disrupted in oncoprotein E2a-Pbx1. Proc Natl Acad Sci U S A, 1997. 94(26): p. 14553-8
- Morgan, R., et al., Identifying HOX paralog groups by the PBX-binding region. Trends Genet, 2000. 16(2): p. 66-7
- Naora, H., et al., A serologically identified tumor antigen encoded by a homeobox gene promotes growth of ovarian epithelial cells. Proceedings of the National Academy of Sciences of the United States of America, 2001. 98(7): p. 4060-4065
- Yamashita, T., et al., Suppression of invasive characteristics by antisense introduction of overexpressed HOX genes in ovarian cancer cells. International Journal of Oncology, 2006. 28(4): p. 931-938
- Naora, H., et al., Aberrant expression of homeobox gene HOXA7 is associated with mullerian-like differentiation of epithelial ovarian tumors and the generation of a specific autologous antibody response. Proceedings of the National Academy of Sciences of the United States of America, 2001. 98(26): p. 15209-15214
- Li, B., et al., HOXA10 is Overexpressed in Human Ovarian Clear Cell Adenocarcinoma and Correlates With Poor Survival. International Journal of Gynecological Cancer, 2009. 19(8): p. 1347-1352
- Fiegl, H., et al., HOXA11 DNA methylation–a novel prognostic biomarker in ovarian cancer. Int J Cancer, 2008. 123(3): p. 725-9
- Morgan, R., Engrailed: complexity and economy of a multi-functional transcription factor. FEBS Lett, 2006. 580(11): p. 2531-3
- Brunet, I., et al., The transcription factor Engrailed-2 guides retinal axons. Nature, 2005. 438(7064): p. 94-8
- Sillitoe, R.V., et al., Engrailed homeobox genes determine the organization of Purkinje cell sagittal stripe gene expression in the adult cerebellum. J Neurosci, 2008. 28(47): p. 12150-62
- Martin, N.L., et al., EN2 is a candidate oncogene in human breast cancer. Oncogene, 2005. 24(46): p. 6890-901
- Morgan, R., et al., Engrailed-2 (EN2): A Tumor Specific Urinary Biomarker for the Early Diagnosis of Prostate Cancer. Clin Cancer Res, 2011
About the Author
After graduating from the University of Cambridge, Richard Morgan held a postdoctoral position at the Hubrecht Laboratory in Utrecht, and then became senior lecturer at St. George’s Medical School, University of London. Richard is currently a principal investigator at the University of Surrey, UK, studying the function of homeodomain-containing transcription factors such as HOX and EN2 in cancer. [email protected]




