Structural genomics, the practical way
Posted: 20 July 2006 | | No comments yet
The Structural Genomics Consortium (SGC) is an internationally funded collaboration with sites in three countries and a three-year goal of solving the 3-dimensional structures of more than 380 human proteins with particular medical relevance, and placing them in the public domain without restrictions. The structures should prove an invaluable resource for research into the proteins’ functions and their use as targets for therapeutic intervention; in this the SGC is a successor to the Human Genome Project (HGP). The SGC has benefited from adopting existing, commercialised robotics, and is subsequently working with vendors to adjust performance with its needs.
The Structural Genomics Consortium (SGC) is an internationally funded collaboration with sites in three countries and a three-year goal of solving the 3-dimensional structures of more than 380 human proteins with particular medical relevance, and placing them in the public domain without restrictions. The structures should prove an invaluable resource for research into the proteins’ functions and their use as targets for therapeutic intervention; in this the SGC is a successor to the Human Genome Project (HGP). The SGC has benefited from adopting existing, commercialised robotics, and is subsequently working with vendors to adjust performance with its needs.
The Structural Genomics Consortium (SGC) is an internationally funded collaboration with sites in three countries and a three-year goal of solving the 3-dimensional structures of more than 380 human proteins with particular medical relevance, and placing them in the public domain without restrictions. The structures should prove an invaluable resource for research into the proteins’ functions and their use as targets for therapeutic intervention; in this the SGC is a successor to the Human Genome Project (HGP). The SGC has benefited from adopting existing, commercialised robotics, and is subsequently working with vendors to adjust performance with its needs.
With only a few nanograms of protein necessary to form a crystal that can yield its full atomic structure, protein crystallography (PX) is by far the most sensitive and accurate technique available for the determination of protein structures. Such a structure typically illuminates much of a protein’s properties and features, so the technique’s popularity is unsurprising. For drug discovery in particular, and most notably for the iterative design of enzyme inhibitors, structures typically suggest an avenue for rationally designing lead compounds that fit the experimental structure, with a number of high-profile successes now on the market (e.g. Relenza®, Celebrex®). Of course, crystals are non-biological artefacts and intrinsically yield static views, but dynamics can be inferred or probed with orthogonal techniques (e.g. NMR).
For all its power, PX suffers the major drawback that it involves a non-trivial, multi-step process with no guarantee that the biological system will crystallise or diffract strongly enough to be solvable: a range of vital systems remain stubbornly inaccessible, in particular membrane proteins and transient multi-component complexes.
Nevertheless, by the end of the 1990s, a series of spectacular technical advances had sped up the procedure enormously, and talk and funding turned to high-throughput crystallography, in particular structural genomics: attempting systematically to solve the structure of all proteins (the proteome) from an organism. Outcomes of the various initiatives – both public and private – have been varied but it did lead to a massive and overdue investment in technology development; in particular, it provided the impetus for the commercialisation of the high-tech instrumentation, to the tremendous benefit of the PX community as a whole.1 Off-the-shelf technology has been the mainstay of PX infrastructure at the SGC-Oxford, where we have sought close collaboration with vendors to optimise its performance.
The Structural Genomics Consortium
The SGC is a four-year, internationally-funded initiative operating from research laboratories at the University of Oxford, UK, the University of Toronto, Canada and the Karolinska Institute in Stockholm. Its primary aim is to solve structures of medically relevant human and malarial protein targets and place them in the public domain, along with experimental details and functional- and binding data generated in the process. Unrestricted access to such information is expected to accelerate research into the respective targets, in particular understanding their roles in disease progression, and hopefully in time to help design new drug leads. The SGC worldwide has set itself the goal to solve over 380 protein structures between July 2004 and June 2007, including proteins associated with cancer, neurological disorders and malaria.
Therapeutic targets: a family approach
Although genes can often be identified as underlying various diseases, it is the proteins they code for that often are the actual biological agents, leading to disease when they malfunction, e.g. deregulated key metabolic or signalling pathways. Through study of the basic biology – and structures – of all components of such a pathway, the range of therapeutic targets can often be broadened considerably. Furthermore, additional structural knowledge of other family members of a target should allow rational design of drugs with greater specificity. Many of the families and their members were known before the HGP; however, availability of the complete sequence, which revealed around 23,000 genes (www.sanger.ac.uk), has significantly increased the possibility of identifying all members of each family, as well their functional and evolutionary relationships.
Here at Oxford ~1000 targets have been selected for study, of which we hope to solve approximately 180 within our three year operational period. The targets cover about 40 structural families, most of which have members that are either involved in disease, or are relevant to other important biological systems. They fall into three broad functional areas:
- Metabolic Enzymes; this covers various enzymes which act upon a large number of structurally and functionally diverse substrates, often associated with metabolic disorders.2,3 Targets include oxidoreductases and 2 oxoglutarate oxygenases and cover a vast range of substrates and functions, from metabolism to signalling and regulation. (Principal Investigator: Udo Opperman)
- Phosphorylation-dependent Signalling; this covers kinases and phosphatases, as well as various regulators of their function. These proteins have been associated with essential cellular process including metabolism, cell migration and communication, and cell cycle events.4,5 Protein Kinases in particular are long-established targets for therapeutic intervention for conditions such as cancer, immunosupression and hypertension. (Principal Investigator: Stefan Knapp)
- Transmembrane Receptor Signalling; this includes a number of integral membrane proteins, but also many of the families of adaptor domains involved in recruiting proteins to signalling events and pathways. These are targets of substantial medical relevance, including ion channels6,7 (e.g. involved in migraine, epilepsy, cystic fibrosis) and G Protein-Coupled Receptors (GPCRs)8 (e.g. hypertension, allergies, congestive heart failure). (Principal Investigator: Declan Doyle)
The family approach has practical benefits, because methodologies and reagents (e.g. ligand libraries) tend to be applicable across a family, thus increasing time- and cost efficiency as well as success rate. Additionally, within families it is not only structure that tends to be conserved but general function too, providing us with a starting point for investigating the mechanism of action for less well characterised targets identified by the HGP.
Finally, it is not clear to what extent specificity really does vary within a family, particularly in the case of the adaptor domains. These are common components of signalling proteins, and are thought to contribute to specificity of protein-protein associations – although alternatively, the subtle molecular differences within a family could be merely evolutionary drift. Structures of many adaptor family members might help address this question.
The Oxford Chapter
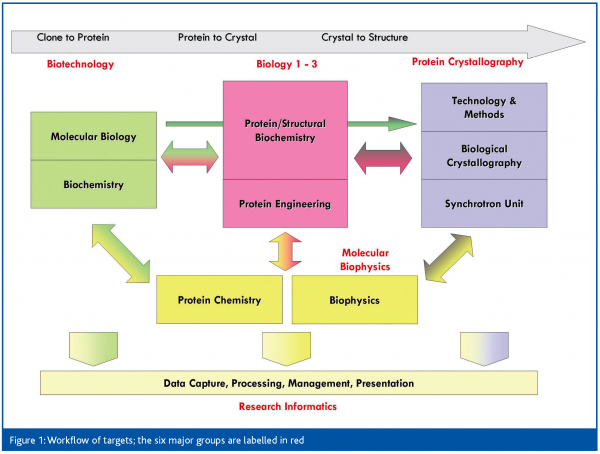
All three SGC sites are structured around the same philosophy: the main focus and investment is in producing pure, well-behaved protein, whereas crystallisation is considered a secondary bottleneck. In Oxford, this means that five of the six primary groups are focused on protein production (Figure 1). The biotechnology group is responsible for clone accrual, initial construct design and initial high-throughput testing of expression and purification of all targets. The three biology groups refine approaches in a target family-specific manner for those that are not readily soluble or crystallised, and convert them to well-behaved proteins. This approach may require construct redesign, limited proteolysis, domain mapping, co-expression, or ligand identification. The biophysics group is relied on for characterisation of protein quality, activity and binding. Successfully purified targets pass into crystallisation, with the protein crystallography (PX) group responsible for crystal optimisation and mounting, data collection, data analysis and structure solution, and finally target deposition. The research informatics group plays a vital role, maintaining the data management infrastructure, and giving bioinformatics support, while the recently established NMR group pursues structures of soluble but non-crystallising proteins.
The SGC thus functions as a close collaboration between the groups – more accurately, between members of the groups; any given project is worked on by only a few scientists across the groups. Projects therefore generally receive careful, individual attention, as they would in a more traditional lab setup. What we deliberately avoided was a ‘pipeline’ approach, which is prone to communication problems and loss of crucial information between stages.
Thanks to the infrastructure and accumulation of reagents and data, this setup allows us scope for orthogonal studies, both on the targets themselves (‘follow-up’), as well as on our methodologies with the aim of improving overall process efficiency. Particularly in the crystallography group, we have an ideal environment for the re-evaluation of established approaches, including the standardisation of difficult crystallographic problems.
Experimental approach
Rather than invest to develop new methodologies, we have deliberately relied on existing, tried-and-tested experimental procedures, focusing instead on how to deploy them efficiently enough to support a high output. Thus for instance, we rely largely on E.coli expression of His-tagged protein in standard vectors; but process at least ten truncation constructs per target, to improve the likelihood of success with this protocol. Likewise, for our initial crystallisation screens, we have relied thus far on commonly-used screening conditions, but reduced their number to 2×96, which simplifies automation logistics, leaving more protein available for subsequent optimisation screens or additional co-crystallisation trials.
At the same time, we cannot sacrifice experimental flexibility: the SGC target list was not selected with convenience in mind, and the targets routinely require more varied protocols than can be supported by standardisation and automation – the alternative is to process even more variations of sample (different constructs/various ligand co-crystallisations; etc.), letting ‘statistical genius’ take care of success. However, that leads to increased costs and lower efficiency, so our challenge is to find the balance between standardised many-sample protocols and experimental best practice. Considering our success rate, the former has worked well enough, but the latter is under constant assessment with the aim of freeing up time and resources for follow-up studies that give meaning to the structures we solve.
The most common systematic obstacles we face are low yields of soluble protein during expression, purification or concentration; moreover, general assays for protein quality do not exist. This poses logistical problems for crystal optimisation by undermining protein reproducibility, especially if frozen storage is not feasible (often in certain classes of protein, e.g. protein kinases). Miniaturisation and automation of subsequent crystallisation are intended to alleviate the situation at least partially, although it is likely to remain as the most frustrating feature of crystallography as a technique.
Leveraging technology
As the SGC was only recently established, we have had the opportunity of using automation tools already on the market, developed on the back of the first wave of Structural Genomics initiatives. This has meant that we have not invested in developing new hardware in-house, as many of the first-phase initiatives were compelled to do. Instead we have been seeking to optimally exploit the hardware and automation already set in place, in particular to support best experimental practices as far as possible.
The infrastructure of the PX group at Oxford comprises:
- Liquid handler for screen preparation (MPII, Perkin Elmer)
- Two nanodrop dispensers for crystallisation setups (mosquito, TTP LabTech)
- Four plate incubators and 2 imagers for automated crystallisation inspections (MinstrelIII, RoboDesign)
- The various tools for crystal mounting and freezing (e.g. Leica microscopes, ALS-style storage pucks);
- X-ray diffractometer with dual port image plate setup (FRE+HTC, Rigaku/MSC);
- Sample changing robot for automated screening of mounted crystals (ACTOR, Rigaku/MSC)
- Regular synchrotron data collection trips at beamline PXII at the Swiss Light Source (as part of a collaboration between protein crystallography groups at the SGC and the SLS).
The majority of crystallisation solutions are mixed by the MPII system, interfaced by a set of home-grown scripts; this applies to both coarse and fine screens, resulting in improved reproducibility as well as savings. Our nanodrop crystallisations are set up by the mosquito systems and stored in and inspected automatically by the MinstrelIII a number of times over a period of two months; images are scored manually with a fast, well-integrated client.
The primary use of our in-house X-ray diffractometer and the attached sample mounting robot is for screening crystals before shipping for synchrotron data collection. This instrumentation is more typically employed for data collection, especially in pharmaceutical research, but in our setup its power allows meaningful ranking of even marginal crystals, thereby hugely increasing beamtime efficiency at the synchrotron. By teaming up with the extremely well-designed SLS PXII synchrotron beamline, we are consistently able to design data collection experiments very carefully, and have an extraordinarily high success rate of synchrotron datasets. To date, 89% of targets collected there have been deposited into the database, and of those, 60% were solved off the first dataset and 60% of datasets are used for final structure determination.
Two technologies in particular have enabled us to leverage small quantities of proteins, and thereby lower cost and effort for protein production. The mosquito, with its ability to accurately pipette nanolitre volumes, allows for much lower protein consumption for crystallisation, and its flexible experiment design is particularly powerful for optimisation screens. Likewise, the very high-performance, well-tuned synchrotron beamline at the SLS allows us realistically to attempt data collection on even very small initial crystals (<50um), allowing for fewer crystal optimisation steps. This has been extremely effective when proteins were difficult to express, or not stable to store long enough for reliable crystal optimisation.
The main objectives for the deployment of our robotics have been to make them effective and accessible; so both experimentally and in throughput, we place high demands. To increase output rather than merely throughput, we have tried to see where experiments can be improved, but invariably this runs into various systems’ limitations. Some examples: For all the increased X-ray power available in modern in-house X-ray diffractometers, including our FRE, the optics and experimental configuration have not been optimised to exploit the power for small crystals. While the MPII has good accuracy and reliability, its software supports only traditional fixed-function applications, and not the flexibility required for mixing more than 100 reagents into varying crystal screens. Through collaboration with the various equipment vendors, we have tried to implement the changes to support our requirements. Since we have limited funding for such activities, we see the possibility of useful changes appearing in future products, and from there into other labs, as the best way of making practical technology contributions to the field.
Of course, the most important aspect of any experiment is data management, and we have spent significant time streamlining and automating data retrieval and transfer from our robotics systems to our home-grown database, BeeHive, allowing the large volumes of data to be managed centrally.
Perhaps the best measure of the usefulness of a deployed technology is internal acceptance, in which case we can claim some success: almost no crystallisations are set up by hand anymore, and almost all plates are submitted for automatic imaging.
Moving forward
The SGC as a whole has generated 16% of all novel human structures in the public database in 2005 (30% so far in 2006) and has contributed more than 250 structures since July 2004, including a number of protein-protein complexes; and it has done so on budget and several months ahead of schedule. Additionally, we have deposited a number of follow-up co-crystal structures to help illuminate function. For the Oxford site, costs per structure are approximately £125k, considered low for human structures. As part of characterising the targets, we are additionally pursuing substrate characterisation, kinetics, screening for binders, and collaborations with external groups to address other questions.
Our family approach is showing results: for at least one family of proteins (the 14-3-3 isoforms), all but two members have been solved by the SGC, and a number of other families are now very well characterised (http://www.sgc.ox.ac.uk/ structures/gallery.html). Knowing the impact of SGC structures on drug discovery is very difficult, but the number and origin of datapack downloads from our Web site indicate great interest; after all, a number of our solved targets had already been solved in industry, but not made publicly available (e.g. FPDS, PIM1 and 11β-HSD1, as evident from coinciding depositions from industry).
In summary, the first years of the SGC have been surprisingly successful, if measured by the original projections and grant conditions. Despite having to maintain a high output of structures, our emphasis has been on the quality of results produced, which was helped by freeing up scientists’ time through the deployment of high-tech systems for crystallisation (e.g. mosquito), analysis and data capture. We have been able to utilise existing technologies in a rather efficient way, helping to optimise the process within the SGC and simultaneously providing extensive feedback to key vendors, indirectly benefiting the community. The next step now is to examine to what extent our particular setup and methodologies are portable; not because they are new, but because they are integrated in a way that clearly works.
We have been pioneering the dissemination of value-added structural information, by publishing on our Web site ‘iSee’ datapacks.9 This is a collection of interactive, annotated views of a structure, hyperlinked to an informative write-up on the structure, and includes details of bioinformatics, materials and methods, and other relevant results (e.g. kinetics). These have aroused great interest in the community, because it is such a more powerful medium for publishing a structure.
Acknowledgements
The Structural Genomics Consortium is a registered charity (number 1097737) funded by the Wellcome Trust, GlaxoSmithKline, Genome Canada, the Canadian Institutes of Health Research, the Ontario Innovation Trust, the Ontario Research and Development Challenge Fund, the Canadian Foundation for Innovation, VINNOVA, The Knut and Alice Wallenberg Foundation, The Swedish Foundation for Strategic Research and Karolinska Institute.


References
- Chandonia JM and Brenner SE. The impact of structural genomics: expectations and outcomes. Science 2006, 311(5759):347-51
- Rinaldo P, Matern D, Bennet MJ. Fatty acid oxidation disorders. Annu Rev. Physiol. 2002, 64:477-502.
- Duax WL, Ghosh D, Pletnev V. Steroid dehydrogenase structures, mechanism of action and disease. Vitam Horm. 2000, 58:121-48.
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Nature. 2002, 298: 1912-1934.
- Cohen P. Protein kinases – the major drug targets of the twenty-first century? Nature Reviews in Drug Discovery. 2002, 1: 309-315.
- Doyle DA, Structural changes during ion channel gating. Trends Neuroscience. 2004, 27:298-302.
- Birch PJ, Dekker LV, James IF, Southan A, Cronk D. Strategies to identify ion channel modulators: current and noval approaches to target neuropathic pain. Drug Discovery Today. 2004, 1: 410-418.
- Klabunde T, Hessler G. Drug design strategies for targeting G-protein coupled receptors. Combichem. 2002, 4:928-944.
- Abagyan R, Lee WH, Raush E, Budagyan L, Totrov M, Sundstrom M, Marsden BD. Disseminating structural genomics data to the public: from a data dump to an animated story. Trends Biochem Sci. 2006




