MS-based clinical proteomics: biomarker discovery in men’s cancer
Posted: 29 October 2010 |
Each year, approximately 10,000 men in the UK die as a result of prostate cancer (PCa) making it the third most common cancer behind lung and breast cancer. Worldwide, more than 670,000 men are diagnosed every year with the disease. Current methods of diagnosis of PCa mainly rely on the detection of elevated prostate-specific antigen (PSA) levels in serum and/or physical examination by a doctor for the detection of an abnormal prostate. PSA is a glycoprotein produced almost exclusively by the epithelial cells of the prostate gland. Its role is not fully understood, although it is known that it forms part of the ejaculate and its function is to solubilise the sperm to give them the mobility to swim. Raised PSA levels in serum are thought to be due to both an increased production of PSA from the proliferated prostate cells, and a diminished architecture of affected cells, allowing an easier distribution of PSA into the wider circulatory system…
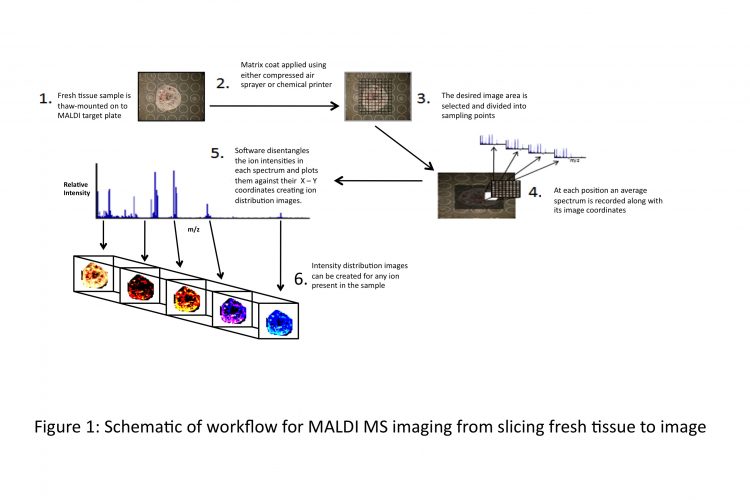

Figure 1 Schematic of workflow for MALDI MS imaging from slicing fresh tissue to image
Each year, approximately 10,000 men in the UK die as a result of prostate cancer (PCa) making it the third most common cancer behind lung and breast cancer. Worldwide, more than 670,000 men are diagnosed every year with the disease1. Current methods of diagnosis of PCa mainly rely on the detection of elevated prostate-specific antigen (PSA) levels in serum and/or physical examination by a doctor for the detection of an abnormal prostate. PSA is a glycoprotein produced almost exclusively by the epithelial cells of the prostate gland2. Its role is not fully understood, although it is known that it forms part of the ejaculate and its function is to solubilise the sperm to give them the mobility to swim. Raised PSA levels in serum are thought to be due to both an increased production of PSA from the proliferated prostate cells, and a diminished architecture of affected cells, allowing an easier distribution of PSA into the wider circulatory system.
There is much debate as to what constitutes a raised level of PSA in the body. The European Association of Urology guidelines released in 2007 state that there is no universally accepted cut-off value between normal and raised PSA levels in the body. The guidelines further state that many studies are using 4ng/ml as the cut-off point between normal and clinically important PSA levels3 – and this threshold increases with age. Aside from a vague universal threshold limit, the PSA test is not specific to prostate cancer alone. Benign prostate hyperplasia (BPH) and prostatitis can also raise PSA levels in men. Equally, there are reported cases of men with below threshold levels of PSA in serum and organ confined prostate cancer. In 2001, Babaian et al found that a significant number of men with PCa (24.5 per cent of a sample size of 51) had serum PSA levels between 2.5 and 4ng/ml4. The problem with lowering the threshold level of PSA as an indicator for both high and low grade prostate cancer was discussed by Yossepowitch in 20083: the detection sensitivity, i.e. number of men correctly diagnosed with PCa, increases at the expense of a concomitant decrease in detection specificity, i.e. increase in the number of men suffering from ‘inconsequential diseases’ such as BPH and undergoing unnecessary biopsies.
The quest for proteomic biomarkers that could serve as more specific identifiers of prostate cancer has been undertaken by a number of research groups using a variety of biological matrices – plasma, serum, urine and prostate secretion. The glycopeptide metalloproteinase inhibitor 1 was recognised by Diamond et al in 20055 as a possible marker of prostate cancer, because it was down regulated in prostate tissue of cancer patients. Another potential marker, named PCa-24 by Zheng et al, was found only to be expressed in patients with prostate cancer6. They used laser microdissection to extract prostate tumour cells and normal/BHP cells from patients undergoing radical prostatectomy. The cells were lysed open and the proteins were selectively analysed using ProteinChip technology and surface-enhanced laser/desorption ionisation time-of-flight mass spectrometry (SELDI TOF MS). They found a protein peak at m/z 24,782.56 ± 107.27 present in 16 of 17 (94 per cent) prostate carcinoma specimens. However, attempts by the group to identify the protein by mass yielded no results6. TB-15 was also reported as a potential biomarker of prostate cancer, as it was found by Hutchinson et al to be up-regulated in patients suffering from prostate or breast cancer7. The same group developed an ELISA for TB-15 and in a recent publication, they report increased specificity from 49 per cent to 64 per cent in a control group of 209 patients. While this study is a welcome addition to the studies that have been carried out examining possible biomarkers for prostate cancer, it has yet to be developed into a specific and sensitive test that out – performs PSA in terms of a robust biomarker differentiating between clinically relevant prostate cancer and neoplasms which might not require further intervention.
Penile cancer is a relatively rare form of cancer, especially in comparison to PCa. It accounts for 1 per 100,000 cancers in Western countries (Europe and North America)8. Approximately 500 men are diagnosed with it each year in the UK9. The fact that it is not as prevalent in society means that there is less research work focused on penile cancer. From literature reviews to date, no proteomic research on penile cancer tissue has been published, so any discoveries using this specific technique could aid in many aspects such as diagnostic biomarker discovery, e.g. for early detection, classification or tumour border identification, or the discovery of new targets for therapeutic drug development. Until recently, the primary treatment for penile cancer was partial/total penectomy or glansectomy8. As with most surgical interventions for cancerous tumours, there is a shift towards more conservative surgical removals and even laser obliteration for the more superficial lesions. Two centimetre borders of normal tissue surrounding the lesion have been the traditional accepted practice for local disease control in penile cancer. Regardless of how small the border is, histopathological examination of the removed tumour is required in all cases to investigate tumour invasion and whether the border cells have been affected by the cancer. Surplus tissue following this pathological examination is made available for this research (study approved by the University of Reading’s Ethics committee and the Berkshire Research Ethics Committee) with the main objective of investigating the distribution of biomolecules in normal, tumorous and border regions of the tissue in situ.
Methods employed for protein biomarker discovery
Proteomics is the large scale exploration into the identification and characterisation of proteins produced in a given organism or tissue. Clinical proteomics is the application of proteomic approaches in clinical research, including the discovery of proteinaceous cancer markers for the purposes of diagnosis and prognosis. In terms of biomarker discovery, proteomics can be used to identify the proteins and their posttranslational modifications, cellular trafficking and even quantitative differences between control and sample groups. Large-scale protein analysis dates back to the 1970s. Laboratories started building up databases of protein expression using some of the newly developed electrophoresis techniques of the time such as 2-dimensional gel electrophoresis (2-DE). Today, 2-DE is still one of the most powerful techniques for protein separation, separating proteins by molecular weight and pI. A major limitation of gel electrophoresis is the fact that it is labour intensive, has low dynamic range and despite recent advances, it still offers only limited resolution for basic proteins or proteins smaller than 10kDa and is highly dependent on protein solubility, making the analysis of hydrophobic proteins such as membrane proteins difficult. Liquid chromatography is also used for the separation of the proteins, and generally, both gel electrophoresis and chromatography precede the mass spectro – metric detection, identification and quantification of the proteins present.
Mass spectrometry applied to protein identification can provide the molecular weight of the protein and can also help unravel the sequence of the protein, especially when techniques like tandem mass spectrometry (MS/MS) are used10. Nonetheless, large-scale proteomic analyses are mainly undertaken at the peptide level after preparative proteolysis, as peptide separation and mass analysis is typically superior for shorter polypeptides. MS/MS coupled with nanoHPLC (such as in MudPIT11) is essentially performing multiple rounds of MS/MS measurements on the many proteolytic products of the proteome that elute from the HPLC system. The approach of multidimensional separation and identification is one of the techniques being utilised in the search for potential biomarkers in this study. A number of search engines have been developed over the recent years (such as MASCOT, SEQUEST and Peptide Prophet) that allow the identification of proteins by matching mass spectrometric data with peptides from primary sequence databases.
A particularly useful ionisation technique in our research is matrix-assisted laser desorption/ionisation mass spectrometry (MALDI), which was pioneered in the late 1980’s12 and is capable of analysing large, non-volatile molecules embedded in a crystalline matrix by employing focused and pulsed laser irradiation. In MALDI, the analyte is co-crystallised with a large molar excess of a matrix compound. The matrix compound used is dependent somewhat on the analyte of interest; however, the general characteristics of a good matrix can be summarised by:
- The matrix compound must sufficiently absorb the laser energy
- The matrix compound must efficiently support the ionisation of the analyte
Commonly used matrices are 2,5-dihydroxy – benzoic acid (DHB), α-cyano-4-hydroxycinnamic acid (CHCA) and sinapinic acid. The MALDI process is dominated by the matrix properties for the sample desorption, and for the ionisation of the analyte through secondary ion transfer reactions. It is characterised by volume disintegration or ablation of the sample rather than desorption of a molecular surface layer, and by proton transfer reactions between the ‘desorbed’ matrix ions and analyte molecules in the created plume. Within a large range of laser pulse durations, it has been shown that MALDI is fluence rather than irradiance dependent with typical fluences around 100-500 J/m2.
In 1997, Richard Caprioli et al13 utilised MALDI to create the first ion distribution map (or MALDI MS image) of tissue. The technique subsequently has been refined and improved upon. The schematic in Figure 1 opposite outlines a workflow of how ion distribution maps are created using a MALDI mass spectrometer. As part of the penile cancer study, MALDI MS imaging will be employed to look at biomolecules present in normal and cancerous cells in situ.
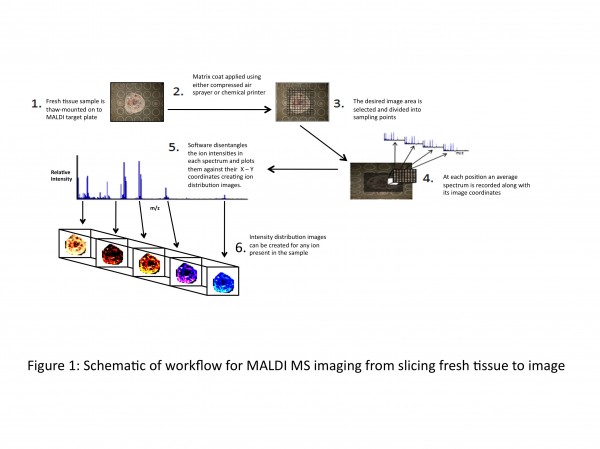

Figure 1 Schematic of workflow for MALDI MS imaging from slicing fresh tissue to image
Samples used for biomarker discovery
Blood plasma and serum are still some of the best sample sources for proteomics-based biomarker investigation, even though their protein complexity and dynamic range create a number of problems. The main problem associated with blood samples is the large range of protein concentrations from Albumin (~50mg/mL) down to low abundant (<1pg/mL) leached tissue proteins such as cardiac myoglobin protein following a heart attack. This makes it difficult to examine all proteins/peptides present in the sample in a one-step analysis. Frequently, the more abundant proteins with higher mass are removed using molecular weight cut-off sieves and filtration to enable the examination of the remaining proteins which are more likely to display biomarker potential.
Urine provides the easiest and least invasive method of sample collection; however, urine samples also display large variability amongst patients. This is in part due to the patient’s consumption of various liquids prior to collection, causing variable dilution or the time of day that collection takes place (first pass urine tends to be more concentrated than urine passed during the day). For sample collection in this study, care is taken to minimise variability, patients are asked to drink one pint of water before attending the clinic, and then standard operating procedures are in place to minimise any variability that may occur by the sample collection.
Royal Berkshire NHS Trust Hospital has an ongoing programme of sample collection from patients attending Urology Procedures Clinic for further investigation into either raised PSA levels or abnormal digital rectal examinations (DRE) given by their GP. The collected samples are urine (pre and post DRE), blood for the plasma extraction and secretion from the tip of the penis during the DRE. These samples are immediately frozen at -80°C and transported to the University of Reading for analysis.
For MALDI MS imaging of penile tissue, method development is being carried out using porcine penile tissue, looking at different matrices and methods of application to achieve optimum detection of ions in the 1,000-10,000 Da mass range as this is the region in which MS-based analytical methods are at their best. A slice of the tissue sample (~10μm thick) is thaw-mounted onto a stainless steel MALDI target plate, and a coat of matrix is applied. There are several methods of applying matrix to the sample. Most frequently used, especially by groups starting out in MALDI MSI, is the compressed-air sprayer (e.g. thin layer chromatography sprayer), applying several coats across the sample but taking care not to over-saturate the sample during the application. There are commercially available chemical printers that ‘print’ the matrix on the tissue, however they are expensive systems. Using in-house developed Perl Imaging Mass Spectrometry Software (PIMSS), the desired image area is then decided upon and divided up into sampling points (the variable distance between laser shots affects the final resolution of the image). The instrument will work its way along the x-axis and down the y-axis, storing an average spectrum from each sampling point along with its image coordinates. Upon completion of MALDI-MS data acquisition, the data is then processed by PIMSS into a readable format for image viewing software such as ‘BioMap’ (free software; http://www.maldi-msi.org)14 or equivalent. This software can disentangle the ion intensities in each spectrum and plot them against their coordinates in order to get a 3-D image (x-position, y-position and the ion intensity) outlining the spatial distribution of any of the ions present in the tissue sample. In terms of biomarker discovery capabilities of these ion densities maps, any disparity in ion intensity in different regions of the tissue (e.g. cancerous cells versus normal cells as identified by a pathologist) can be investigated for the possibility of up or down-regulation of any biomolecules or other differences that may be translated into a viable discrimination tool for ‘abnormal’ cells against ‘normal’ cells.
It is hoped that novel biomarkers will be found with these new MS-based analytical methods that can provide an indiscriminate view of the molecular landscape of cancer at high sensitivity. To achieve this, advanced proteomic techniques (including rigorous sample collection) are applied. Some of these have already been applied to the more common prostate cancer15,16 while none of them have so far been used for the discovery of penile cancer biomarker.
References
1. Cancer Research UK. Cancer research Statistics 2009 [cited 2009 03/11/2009]; Web based statistics. Available from: http://info.cancerresearchuk.org/cancerstats/ types/prostate/index.htm?script=true
2. Barry, M.J., Clinical practice. Prostate-specificantigen testing for early diagnosis of prostate cancer. N Engl J Med, 2001. 344(18): p. 1373-7
3. Cancer Research UK. Cancer research Statistics 2009 [cited 2009 03/11/2009]; Web based statistics. Available from: http://info.cancerresearchuk.org/cancerstats/ types/prostate/index.htm?script=true
4. Barry, M.J., Clinical practice. Prostatespecific- antigen testing for early diagnosis of prostate cancer. N Engl J Med, 2001. 344(18): p. 1373-7
5. Yossepowitch, O., Prostate cancer in men with serum PSA of
6. Babaian, R.J., et al., The incidence of prostate cancer in a screening population with a serum prostate specific antigen between 2.5 and 4.0 ng/ml: relation to biopsy strategy. J Urol, 2001. 165(3): p. 757-60
7. Liu, A.Y., et al., Analysis of prostate cancer by proteomics using tissue specimens. J Urol, 2005. 173(1): p. 73-8
8. Zheng, Y., et al., Prostate carcinoma tissue proteomics for biomarker discovery. Cancer, 2003. 98(12): p. 2576-2582
9. Hutchinson, L.M., et al., Use of thymosin β15 as a urinary biomarker in human prostate cancer. The Prostate, 2005. 64(2): p. 116-127
10. Salvioni, R., et al., Penile cancer. Urol Oncol, 2009. 27(6): p. 677-85
11. Cancer Research UK. Risks and causes of penile cancer. [Web Page ] 2009 [cited 2009 23/11/2009]; Available from: http://www.cancerhelp.org.uk/type/penilecancer/ about/risks-and-causes-of-penile-cancer
12. de Hoffmann. Edmond and Stroobant. Vincent, Mass Spectrometry Principles and Applications 3rd ed. 2009, Chichester: John Wiley & Sons Ltd. 479
13. Washburn, M.P., D. Wolters, and J.R. Yates, Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotech, 2001. 19(3): p. 242-247
14. Karas, M. and F. Hillenkamp, Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal Chem, 1988. 60(20): p. 2299-301
15. Caprioli, R.M., T.B. Farmer and J. Gile, Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem, 1997. 69(23): p. 4751-60
16. Stoeckli, M., et al., Imaging mass spectrometry: a new technology for the analysis of protein expression in mammalian tissues. Nat Med, 2001. 7(4): p. 493-6
17. Rehman, I., et al., Proteomic analysis of voided urine after prostatic massage from patients with prostate cancer: a pilot study. Urology, 2004. 64(6): p. 1238-43
18. Schwamborn, K., et al., Identifying prostate carcinoma by MALDI-Imaging. Int J Mol Med, 2007. 20(2): p. 155-159
About the Authors
Brian Flatley
Brian completed a B.Sc. (Hons.) in Pharmaceutical & Forensic Analysis at Limerick Institute of Technology in 2008. Following the completion of his undergraduate degree, he worked for the Irish State Laboratory’s toxicology department. Brian started in the University of Reading under the joint supervision of Professor Cramer (University of Reading) and Mr Peter Malone (Harold Hopkins Department of Urology, Royal Berkshire NHS Foundation Trust Hospital) in October 2009. The remit of his study is “Mass spectrometry based biomarker discovery in men’s cancer.” The main diseases that the research is focused on are prostate and penile cancer.
Rainer Cramer
Rainer is an experimental physicist and chemist with a strong background in biological mass spectrometry and (bio)analytical and protein/peptide chemistry. He is Chemistry Professor for Bioanalytical Sciences and Mass Spectrometry and Head of Mass Spectrometry and Proteomics in the BioCentre at the University of Reading. His previous positions were at the Ludwig Institute for Cancer Research as Assistant Member and Head of the Bioanalytical Chemistry and Mass Spectrometry group. He was also affiliated with UCL, London, where he was Reader in Biological Mass Spectrometry. He obtained his PhD in Professor Hillenkamp’s laboratory, the birthplace of MALDI-MS.





