Genome-wide High Content Analysis of cellular pathways
Posted: 23 January 2008 | Jeremy C. Simpson, Cell Biology and Biophysics Unit, European Molecular Biology Laboratory (EMBL) | No comments yet
Creating the molecular tools to combat human disease and infection remains the cornerstone activity of the pharmaceutical industry. The methodologies employed to discover new drugs has continually evolved as new biological techniques have emerged1; nevertheless the development of each novel compound is still only realised after many years of careful research, and a detailed analysis […]
Creating the molecular tools to combat human disease and infection remains the cornerstone activity of the pharmaceutical industry. The methodologies employed to discover new drugs has continually evolved as new biological techniques have emerged1; nevertheless the development of each novel compound is still only realised after many years of careful research, and a detailed analysis of its specific target.
These targets can potentially be any class of protein, and their selection is dependent on gathering sufficient high quality data about the pathway and reactions in which they are involved. The cataloguing of genes, and therefore proteins, that has come from genome-wide sequencing efforts now provides a resource from which new drug targets can be identified; however, the challenge is first to assign each of these genes to a particular pathway and function. One approach that can help achieve this is the use of microscopy-based assays in cultured cells. Cell-based assays are now well established in chemical compound screening to identify molecules affecting particular pathways2, but this does not globally address gene function. However, if cell-based assays are combined with protein over-expression or down-regulation approaches, gene function can be probed. Subsequent high content analysis (HCA) of the cells after such treatments can then provide quantitative and contextual information about the role of every gene in a particular pathway. This review will highlight some of the issues faced by researchers now employing this strategy.
The importance of subcellular architecture
Gene function may be ascertained using a variety of experimental approaches, including biochemical techniques, in vitro assays, and genetics. Although each method has its own particular strengths, none of these consider the cell as an individual entity, and as such do not take account of the exquisite spatial arrangement of its subcellular organelles, that in turn permit the partitioning of biochemical reactions and pathways. Conversely, high content screening (HCS) and analysis relies on the morphological characteristics of cells, and in particular the fact that changes to cell shape, subcellular structures, or even individual markers can occur after drug treatment or gene perturbation, and that these changes can be detected and quantified by automated microscopy3.
One important step on the road to determining gene function is the identification of the subcellular compartment to which the corresponding protein localises. However, it is unlikely that a single experimental approach will be able to localise any complete mammalian proteome, but rather combined efforts using proteomics, antibody collections, and expression of epitope- or fluorescently-tagged open reading frames (ORFs), will together make attaining this goal a realistic possibility in the near future4. For HCS to have a greater impact in determining the localisation of entire proteomes, two principal requirements must be fulfilled: Firstly, comprehensive collections of tagged ORFs need to be generated and expressed in cells. Many individual clone collections now contain in excess 10,000 ORFs5-7, however due to the splicing complexity found in mammalian organisms it is unlikely that even the pool of these collections will contain a complete proteome.
Secondly, computational tools must accurately annotate localisation. Automated recognition of subcellular patterns has recently seen dramatic advances8, but a lack of standardisation of transfection techniques, cell lines, and image acquisition parameters between laboratories means that some problems still need to be overcome. Indeed, even the expression of fluorescently tagged ORFs encoding different proteins that all localise to the same subcellular compartment can still result in a diverse morphological appearance of the organelle (see Figure 1 for an example), and therefore the analysis software needs to be sufficiently robust to compensate for this.
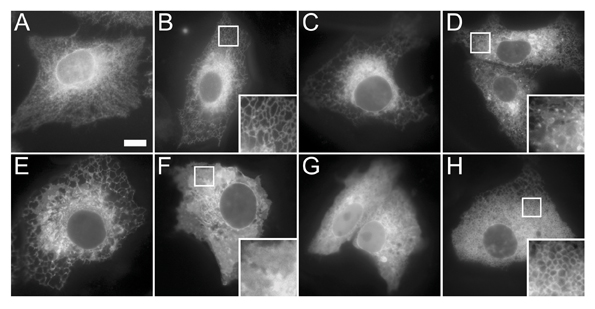

Figure 1: Diverse morphologies of the endoplasmic reticulum. (A) Antibody staining of the ER chaperone calnexin in Vero cells showing a typical reticular distribution throughout the entire cell. (B-H) GFP-tagged open reading frames expressed in Vero cells for 24 hours and then imaged in the living cells. All of the expressed proteins localise to ER membranes, however the morphology of this organelle appears very different depending on the protein expressed. The proteins in panels B-D show a predominantly reticular pattern, although in panel D a number of distinct punctate structures can also be seen. The proteins in panels E and F show a more sheet-like appearance. The protein in panel G also displays a significant amount of soluble cytoplasmic signal, and the protein in panel H causes a abnormal vesiculation of the ER.
Systematic perturbation of gene function
A parallel method to systematically probe gene function is to combine the modulation of protein expression level in vivo with an assay that gives a specific and quantifiable readout. The development of automated microscopy platforms means that not only can this approach be performed in intact and living cells, but that it can be carried out on a large or even genome-wide scale. ‘Gain-of-function’ by over-expression has long been the preferred choice for modulating the level of gene action, however the relatively recent advent of RNAi interference (RNAi) methodologies in cultured cells means that systematic ‘loss-of-function’ studies are now also feasible. As discussed above, due to the incomplete sets of ORFs currently available, over-expression studies cannot yet be comprehensive and, furthermore, care must be taken to avoid artefacts as a consequence of poorly controlled expression levels. Nevertheless, the expression of fluorescently tagged proteins in living cells provides the advantage that each protein under study can be directly monitored in real time, and its relative expression quantitatively correlated to any phenotype observed. Our own laboratory has successfully used this approach to test the effect of over-expression of more than 100 proteins on constitutive secretion, resulting in the identification of 20 proteins that either inhibited secretion or caused an aberrant Golgi complex morphology9. Using RNAi as a method to analyse gene function on a global scale is now becoming a possibility. Once again, it is important to note that this opportunity has only arisen as a direct result of genome sequencing projects, and that as the accuracy of genome annotation continues to improve, so will the RNAi reagents themselves. Although there are currently more examples of successful large-scale RNAi-based screens carried out in cells from model organisms such as Drosophila melangaster10-12 – largely due to the fact that RNAi libraries became more rapidly available for such organisms – we are now beginning to see examples of such screens being undertaken in human cell lines. Of note is a complete analysis of kinase involvement in endocytosis pathways carried out in HeLa cells13. This study was particularly striking in that by using two viruses – vesicular stomatitis virus (VSV) and Simian virus 40 (SV40) – Pelkmans and co-workers were able to probe two endocytosis pathways with a common set of RNAi reagents, and thereby identify factors that were both common and distinct between the pathways. More recently, a complete genome-wide analysis of cell division in HeLa cells has also been reported14. In this work the primary screen utilised propidium iodide staining of chromatin 72 hours after transfection, with RNAi reagents followed by analysis of the DNA content. The 2,146 hits obtained were then taken into validation screens, and then the remaining 1,351 positives into more detailed secondary screens employing measurements of mitotic index and cell size. Finally, video microscopy was used to determine the specific mitotic defect in a number of the interesting candidates. Curiously, when the authors compared their final hit lists to those hits obtained in an independent, albeit similar screen for cell cycle regulators15, an overlap of only 10% between the data sets was observed. It is important to note that although these two screens were carried out in different cell lines (HeLa and U2-OS), this alone cannot account for this disparity. Improved automated microscopy now enables HCS time-lapse experiments to be carried out over several days and several hundred genes to be analysed in parallel16, and therefore the application of this type of screening regime should improve our understanding of cell cycle and cell division pathways. Similarly, many other cellular pathways and processes are now likely to be studied on a global scale. We ourselves have developed an integrated strategy to search for novel players involved in protein secretion17, and assays to study other pathways or organelles will surely follow.
HCS assay design and implementation
Despite the precedents that have now been established for using large-scale assays to determine gene function, actually designing and embarking on a screening regime is not a task that should be lightly undertaken. Although detailed guidelines relating to assay design are beyond the scope of this article, and have been covered recently in an excellent review by Echeverri and Perrimon18, a few salient features are worth pointing out. The first of these is that it is essential to have a validated and robust assay that is proven to work on a small scale (no more than tens of genes), prior to beginning any high-throughput experiments. Another key issue relevant to RNAi screening is both the type of library to be used, and the methodology with which to deliver it into cells. RNAi reagents are now available in various forms, including chemically synthesised small interfering RNAs (siRNAs), vector-based short hairpin RNAs (shRNAs), and endoribonuclease prepared siRNAs (esiRNAs), with each library having its own advantages and disadvantages depending on the application19. Similarly, there are now choices to be made in terms of how to deliver these reagents into cells. Possibilities include; viral vector systems, conventional liquid-based transfection, and reverse transfection. This latter methodology is particularly attractive, as compared with liquid-based transfection, only relatively tiny amounts of RNAi reagents are needed for printing on to glass slides or coverglass chambers. These arrays can be printed at high density, with many replicates prepared in parallel, and they can be stored desiccated for several months without loss of activity20.
Data analysis also presents important issues to successful HCS implementation. One principal advantage of HCS over biochemical-based assays is that potentially an infinite number of parameters can be extracted from an image; therefore it is important that these data are used appropriately. Images need to be processed in a consistent manner (for example, including background subtraction and thresholding) and analysis parameters need to be chosen that truly reflect what can be seen in the cells (see example in Figure 2). A huge variety of commercial software packages to do this are now available. Similarly, having converted image data into numerical data, appropriate statistical methods should also be employed21.
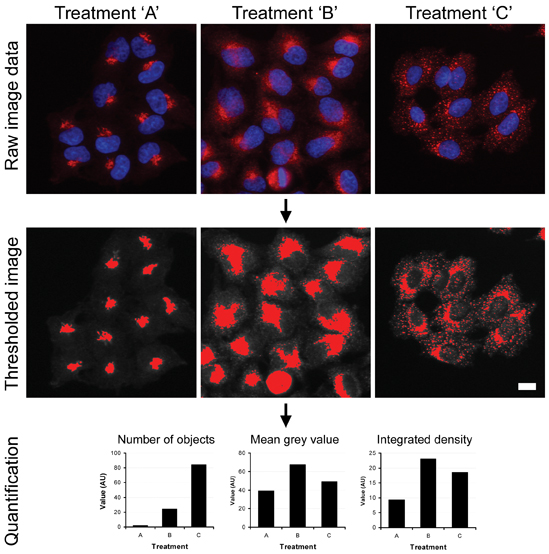

Figure 2: High content analysis of complex subcellular structures. HeLa cells were fixed and then immunostained with antibodies against the Golgi marker GM130 (red channel in upper panel) following treatment with control buffer (treatment A) or two different chemical reagents known to perturb Golgi morphology (treatments B and C). The images were then background-subtracted and then thresholded to identify structures of interest. Multi-parameter analysis was then carried out on these structures on a cell-by-cell basis using ImageJ software.
Finally it is also worth bearing in mind that any HCS regime to assess gene relevance to a particular pathway only represents the first step in analysis. Primary screening data needs to be supported with validation screening, ideally using independent reagents (additional RNAi sequences from alternative manufacturers or sources), and that candidate genes successfully coming through this stage require more detailed examination in secondary assays. Off-target effects and false positives remain a problem in this field22, and so it is imperative that large-scale data sets are integrated with one another so that greater confidence in the results is gained.
Conclusions
Genome-wide HCS approaches have now become a realistic mechanism to dissect gene function in both commercial and academic laboratories. The reagents to carry out these experiments are becoming more accessible, and the experience that has already been gained in this relatively young field of biological research needs to be passed on to those who are just embarking on screening regimes. HCS is a means to truly reap the information gained from genome sequencing projects, such that genes can be comprehensively married to biological pathways. This will accelerate our understanding of cell function, in turn enhancing drug discovery efforts.
References
- Drews J: Drug discovery: A historical perspective. Science. 2000, 287:1960-4.
- Korn K, Krausz E: Cell-based high-content screening of small-molecule libraries. Curr Opin Chem Biol. 2007, 11:503-10.
- Pepperkok R, Ellenberg J: High-throughput fluorescence microscopy for systems biology. Nat Rev Mol Cell Biol. 2006, 7(9):690-6.
- Simpson JC, Pepperkok R: The subcellular localization of the mammalian proteome comes a fraction closer. Genome Biol. 2006, 7(6):213.
- Strausberg RL, et al.: Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci USA. 2002, 99(26):16899-903.
- Lamesch P, et al.: hORFeome v3.1: A resource of human open reading frames representing over 10,000 human genes. Genomics. 2007, 89:307-15.
- Bechtel S, et al.: The full-ORF clone resource of the German cDNA consortium. BMC Genomics. 2007, 8(1):399.
- Glory E, Murphy RF: Automated subcellular location determination and high-throughput microscopy. Dev Cell. 2007, 12:7-16.
- Starkuviene V, Liebel U, Simpson JC, Erfle H, Poustka A, Wiemann S, Pepperkok R: High-content screening microscopy identifies novel proteins with a putative role in secretory membrane traffic. Genome Res. 2004, 14:1948-1956.
- Boutros M, et al.: Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004, 303(5659:832-5.
- Bard F, et al.: Functional genomics reveals genes involved in protein secretion and Golgi organization. Nature. 2006, 439:604-7.
- Yi CH, Sogah DK, Boyce M, Degterev A, Christofferson DE, Yuan J: A genome-wide RNAi screen reveals multiple regulators of caspase activation. J Cell Biol. 2007, 179(4):619-26.
- Pelkmans L, Fava E, Grabner H, Hannus M, Habermann B, Krausz E, Zerial M: Genome-wide analysis of human kinases in clathrin- and caveolae/raft-mediated endocytosis. Nature. 2005, 436:78-86.
- Kittler R et al.: Genome-scale RNAi profiling of cell division in human tissue culture cells. Nat Cell Biol. 2007, 9(12):1401-12.
- Mukherji M, et al.: Genome-wide functional analysis of human cell-cycle regulators. Proc Natl Acad Sci USA. 2006, 103(40):14819-24.
- Neumann B, Held M, Liebel U, Erfle H, Rogers P, Pepperkok R, Ellenberg J: High-throughput RNAi screening by time-lapse imaging of live human cells. Nat Methods 2006, 3:385-390.
- Simpson JC, Cetin C, Erfle H, Joggerst B, Liebel U, Ellenberg J, Pepperkok R: An RNAi screening platform to identify secretion machinery in mammalian cells. J Biotech. 2007, 129(2):352-65.
- Echeverri CJ, Perrimon N: High-throughput RNAi screening in cultured cells: a user’s guide. Nat Rev Genetics. 2006, 7:373-84.
- Clark J, Ding S: Generation of RNAi libraries for high-throughput screens. J Biomed Biotech. 2006, 2006:1-7.
- Erfle H, Neumann B, Liebel U, Rogers P, Held M, Walter T, Ellenberg J, Pepperkok R: Reverse transfection on cell arrays for high content screening microscopy. Nat Protocols. 2007, 2(2):392-9.
- Ainscow E: Statistical techniques for handling high content screening data. Eur Pharmaceutical Rev. 2007, 5:30-8.
- Echeverri CJ, et al.: Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat Methods. 2006, 3(10):777-9.
Jeremy C. Simpson
Cell Biology and Biophysics Unit, European Molecular Biology Laboratory (EMBL)
Jeremy Simpson obtained a BSc degree in Microbiology and Microbial Technology from the University of Warwick, UK. He also carried out his PhD work in the same university, in the laboratory of Mike Lord and Lynne Roberts, working on toxin trafficking in mammalian cells. After brief post-doctoral work in London (ICRF) and San Diego (Scripps), he was awarded an EMBO Long Term Fellowship enabling him to move to the newly established laboratory of Rainer Pepperkok at the EMBL in Heidelberg, Germany. Since 2001 he has been a staff member at EMBL, developing high-throughput methods to study membrane traffic. In Spring 2008 he takes up a position as Professor of Cell Biology at University College Dublin, Ireland.
Issue
Related topics
Related organisations
Cellular Pathways, European Molecular Biology Laboratory (EMBL)




