Real-time polymerase chain reaction – towards a more reliable, accurate and relevant assay
Posted: 3 December 2008 | Stephen A Bustin BA(Mod) PhD, Professor of Molecular Science, Queen Mary University of London | No comments yet
The fluorescence-based quantitative real-time polymerase chain reaction (qPCR)1,2,3 has become firmly established as the preferred technology for the detection and quantification of nucleic acids in molecular diagnostics, life sciences, agriculture and medicine4,5.
The fluorescence-based quantitative real-time polymerase chain reaction (qPCR)1,2,3 has become firmly established as the preferred technology for the detection and quantification of nucleic acids in molecular diagnostics, life sciences, agriculture and medicine4,5.
The fluorescence-based quantitative real-time polymerase chain reaction (qPCR)1-3 has become firmly established as the preferred technology for the detection and quantification of nucleic acids in molecular diagnostics, life sciences, agriculture and medicine4,5. The combination of conceptual and practical simplicity, large dynamic range of linear quantification, speed, sensitivity and specificity has made it the yardstick for nucleic acid quantification not just in basic research, but has engendered numerous uses ranging from basic research through diagnostic and forensic application to treatment monitoring in a clinical setting6-11. Despite its ubiquity, the coming-of-age of this technology has been hampered by significant biological as well as technical issues that frequently combine to obfuscate the interpretation of qPCR data. Biological issues have been discussed elsewhere9,10,12,13; nevertheless, it is essential to always keep biological relevance in mind when interpreting results, especially when they relate to patient prognosis or drug monitoring. Technical issues are centered on experimental design, data handling, analysis and reporting and whilst peripherally acknowledged, are frequently not adequately addressed4,14,15,16,17. This insouciant attitude has had major repercussions in the public health domain. A combination of flawed use of qPCR technology, specious data analysis and flawed interpretation resulted in the detection of RNA measles virus in the intestines of children with developmental disorders18. These data were central to the widespread speculation linking the measles, mumps and rubella (MMR) vaccine with the development of autism. This, in turn, has caused untold distress to thousands of parents, resulted in a dramatic decline in MMR vaccination in a number of countries and was the subject of major class actions in both the UK and the US. At last year’s trial at the vaccine court in Washington DC the reliability of the qPCR data was seriously challenged, with DNA contamination shown to be the most likely cause of most, if not all, positive results19. Furthermore, the data could never be independently reproduced20,21,22. Nevertheless, some lingering doubt continued to remain until recently, when a paper was published that concludes that there is no evidence for an association between persistent measles virus RNA in the gut and autism23. Astonishingly, this publication includes the two main authors of the original paper, and despite publishing evidence that contradicts their own, they have not retracted their original paper. Another example of problems associated with qPCR technology concerns a Science magazine “breakthrough of the year 2005”24, which has had to be retracted because of the consequences of poor qPCR assay execution. Consequently it has been clear for a while that not only are stringent quality control checkpoints at each stage of the experimental workflow indispensable components of a well-designed qPCR experiment, but that these checkpoints must be verifiable. This has resulted in a number of recent initiatives, all aimed at improving the reliability of qPCR-derived data and the transparency of data reporting. 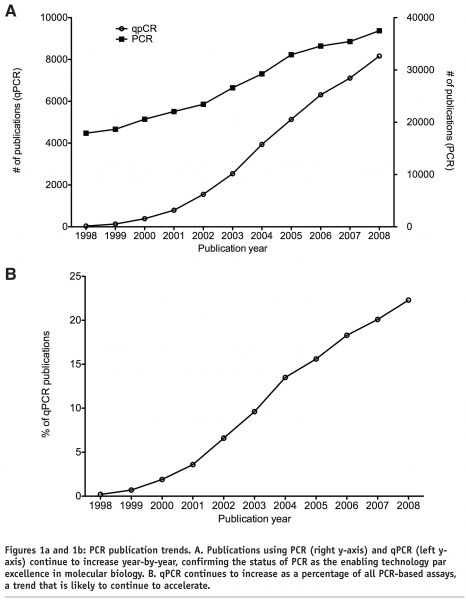

Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE)
Many publications utilising qPCR technology barely provide sufficient information to permit the reader to evaluate the validity of any conclusions derived from the quantitative qPCR data. For example, it is universally accepted that RNA quality assessment is essential for reliable quantification of cellular mRNA using RT-qPCR assays25-29. Yet a brief perusal of 50 BMC open access publications from January to April 2008 reveals that 31 (62%) do not even mention RNA quality, and a further five (10%) describe A260:280 ratios that are generally accepted as inadequate for accurate quantification. Similarly, a very high percentage of papers continue to normalise gene of interest copy numbers against single reference genes, despite the definitive demonstration that this general approach is invalid30-33 and the ready availability of several methods allowing the selection of appropriate sets of reference genes34,35,36. Other common omissions concern information on sample handling and storage, primers and probes selection, details of the reverse transcription step, efficiency of the PCR reaction, inclusion of controls and data analysis. A summary of the minimum information required to reproduce a rt-qPCR assay is provided in Figure 2. 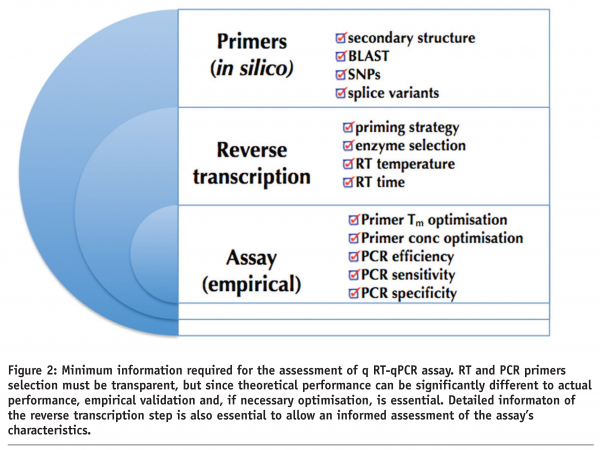


Real-time PCR Data Markup Language (RDML)
In the qPCR Stone Age, i.e. around ten years ago, there was a grand choice of two real-time instruments. Characteristically, even then they used incompatible formats (rotor vs. 96-well plate), and quantification schemes (baseline/threshold vs. the second derivative maximum). The former determines the quantification cycle Cq by drawing a line parallel to the x-axis of the amplification plot and noting the cycle fraction where it crosses the log-linear phase of the amplification plot. Unfortunately, this makes the precise positioning of the threshold line entirely subjective. The latter method calculates the point of maximal increase of fluorescence within the log-linear phase by determining the second derivative maxima of the amplification curves. The software calculates at which cycle number this point is reached. Both methods remain in common use. To-day there is a great number of instruments available, all utilising different formats, technologies and software. This proliferation is coupled to the appearance of instruments capable of high throughput: several (Lightcycler 480, BioRad CFX384, Applied Biosystems 7900HT) have 384 well blocks and Biotrove’s OpenArray and Fluidigm’s BioMark technologies permit the analysis of approximately 10,000 reactions simultaneously. Three important consequences of this are that (1) assays are being carried out on a multitude of platforms giving potentially different results, (2) the number of specialised applications is continuously increasing, and (3) the number of samples analysed during each run is escalating. Whereas initially most qPCR studies quantified the expression of a handful of target genes in response to, say, drug administration, the introduction of these high throughput systems allows the quantification of numerous genes by qPCR. Consequently qPCR experiments are beginning to match the assessment of complex biological phenomena in the context of high dimensional gene expression profiling. This proliferation of platforms has resulted in a concomitant increase of instrument-specific software used to generate qPCR data. Since manufacturers do not consider end-user convenience when designing their software, data formats are incompatible. This not only limits data import and export, but data handling and quality control become non-transparent45. Every manufacturer saves their run files in proprietary formats and, whilst information can be exported in various file formats (.CSV, .TXT, .XLS), all have different layout and data field terminologies. This creates unnecessary problems for users wanting to exchange data between instrument-specific software packages and analysis tools or for collaborators wanting to share qPCR data between different laboratories. The result is a “Tower of Babel” syndrome, where every instrument speaks a different language, which serves to obscure, confuse and limit data exchange. A possible solution to this was unveiled in 2005, when a universal XML-based data format initiative for the exchange and publication of qPCR data was proposed at the Freising qPCR Symposium (www.wzw.tum.de/gene-quantification/qpcr2005/pub/). Its aim was to encourage the adoption of a universal data format for real-time PCR data, named RDML (Real-time PCR Data Markup Language). This initiative was followed by the launch of a RDML-website in 2006 and the inauguration of the RDML consortium (www.rdml.org) in 200844,46. When implemented, the universal data format will provide sufficient information to allow reviewers and reader to understand the qPCR experimental setup, re-analyse the data and interpret the results. It will be independent of computer hardware, operating system or available software package, and sufficiently flexible to allow future additions of additional information. The main advantage of a common universal format would be the ease with which raw annotated data could be supplied to manuscript reviewers and readers, collaborators and databases. The main disadvantages are that there is currently no agreed standard for what information should be included, how precisely the information should be handled, whether and how instrument manufacturers would modify their software and whether researchers would be willing to see their raw data re-analysed and re-interpreted. Nevertheless, it does not require much imagination to predict that some implementation of a common data format will become more and more essential and that such a step would be a significant extension of the power of qPCR technology.
Data analysis and management programs
Although the analysis of qPCR data has been described as having reached a mature stage of development47, the publication of disparate and often contradictory analysis methods suggests that there is little consensus with respect to data analysis and probably even less consistency of data management. Consequently, the coordination and efficient management of experimental data, especially with respect to their statistical evaluation, represent a critical additional, albeit poorly addressed, challenge. Furthermore, the increasing trend towards complex, high throughput qPCR experiments places robust data analysis and management tools at the heart of the drive towards creating a sophisticated data analysis environment yielding reliable and reproducible qPCR data. The appreciation that PCR efficiency was variable, and hence a key parameter for establishing reliable PCR assays, has resulted in the publication of numerous methods, both linear and non-linear, for calculating optimal (i.e. most accurate) PCR efficiency48-59. Many of the concepts and algorithms developed in these publications lie at the core of the numerous software packages that have been developed for data analysis. One downside of this diversity, incidentally, is that this diversity may lead to variable or even inaccurate results60. Although instrument-specific software has become more flexible and powerful, its development has not kept pace with the major technological advances incorporated into the instruments. All supplied software can extract Cq information from recorded fluorescence measurements, allowing the user to display basic amplification plots, together with threshold lines and melt curves. However, beyond the construction of standard curves, the calculation of sample copy numbers, means and standard deviation for replicates, both their functionality and sophistication are severely limited. Raw data processing is only just beginning to be incorporated by instrument manufacturers: for example BioRad’s CFX software allows limited gene expression analysis by providing options for calculation of differences in a target’s concentration between samples, showing them either as normalised expression (ΔΔCq) or relative quantity (ΔCq). The need for better experimental organisation and more powerful, statistically reliable data analysis has led to the development of a number of software tools that are designed to standardise, simplify and make qPCR data management and analysis more transparent. One such tool is PREXCEL-Q61,62, which is unique in that it addresses the labour- and time-intensive set-up and optimisation steps associated with the introduction of a new qPCR assay. It provides a comprehensive set of Excel-based templates that permit rapid calculation of reagent volumes, e.g. for nuclease treatments or RT and qPCR reactions and generates ready-to use protocol printouts. Especially useful is the feature that allows the researcher to identify appropriate sample dilutions that result in maximum amplification efficiency. Another category of analysis tool aims to address every step of a qPCR assay by incorporating appropriate algorithms for quantification, statistical tests, error propagation, inclusion of data quality control etc. Following the import of raw Cq data, they may perform quality control and outlier detection, examine the correlation between biological replicates, select the optimal combination of endogenous controls for normalisation based on stability algorithms and compute fold-change and significance results for differential expression analysis. An early example is the modular Q-Gene63, with more recent programs becoming increasingly sophisticated, comprehensive and user-friendly, e.g. qPCR-DAMS64, DATAN GenEx and qBasePlus45. Use of such software not only speeds up the analysis of raw RT-qPCR data, but also helps improve experimental accuracy by implementing more rigorous analytical workflows. They offer a range of sophisticated multivariate analyses, incorporate algorithms for identifying optimal normalisation genes, e.g. geNorm34 or NormFinder36 and use inter-run calibration methods that allow samples analysed in different runs to be compared against each other. For users of Applied Biosystems instruments, Integromics’ StatMiner adds data mining capability to enhance qPCR data analysis by using functional annotations from public databases.
Conclusion
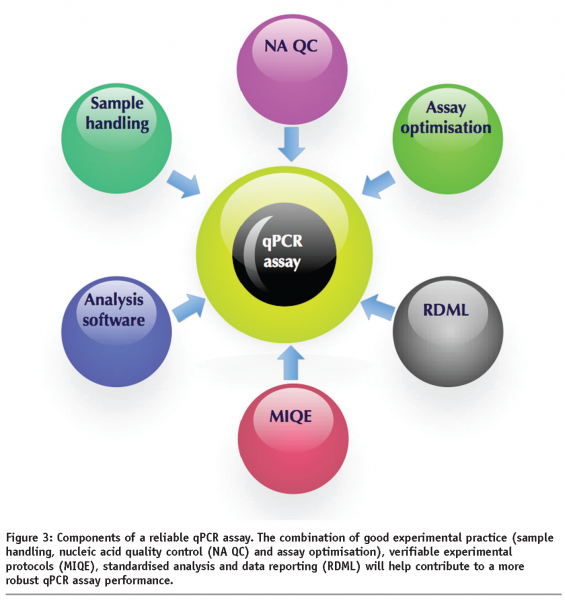
qPCR has been passing through a “cowboy” phase that is characterised by a technological free-for-all in terms of methodology, protocols, data analysis and interpretation and consensus on the amount of information required for publication. However, qPCR has been around for 16 years and is no longer a novel technique. Consequently it is high time for the technology to enter a more consolidated period that will yield verifiable technically reliable, as well as biologically, meaningful data. A combination of appropriate experimental design and an acceptance of the three initiatives discussed above would constitute a significant step towards this goal and will, it is hoped, allow qPCR technology to fulfill its immense promise.
References
- R. Higuchi, G. Dollinger, P.S. Walsh, and R. Griffith, Simultaneous amplification and detection of specific DNA sequences. Biotechnology (N.Y.) 10 (1992) 413-417.
- R. Higuchi, C. Fockler, G. Dollinger, and R. Watson, Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology (NY) 11 (1993) 1026-1030.
- C.T. Wittwer, M.G. Herrmann, A.A. Moss, and R.P. Rasmussen, Continuous fluorescence monitoring of rapid cycle DNA amplification. Biotechniques 22 (1997) 130-138.
- S.A. Bustin, Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 25 (2000) 169-193.
- M. Kubista, J.M. Andrade, M. Bengtsson, A. Forootan, J. Jonak, K. Lind, R. Sindelka, R. Sjoback, B. Sjogreen, L. Strombom, A. Stahlberg, and N. Zoric, The real-time polymerase chain reaction. Mol Aspects Med 27 (2006) 95-125.
- P.S. Bernard, and C.T. Wittwer, Real-time PCR technology for cancer diagnostics. Clin Chem 48 (2002) 1178-85.
- I.M. Mackay, K.E. Arden, and A. Nitsche, Real-time PCR in virology. Nucleic Acids Res 30 (2002) 1292-305.
- I.M. Mackay, Real-time PCR in the microbiology laboratory. Clin Microbiol Infect 10 (2004) 190-212.
- S.A. Bustin, and R. Mueller, Real-time reverse transcription PCR (qRT-PCR) and its potential use in clinical diagnosis. Clin Sci (Lond) 109 (2005) 365-79.
- S.A. Bustin, and R. Mueller, Real-time reverse transcription PCR and the detection of occult disease in colorectal cancer. Mol Aspects Med 27 (2006) 192-223.
- R.J. van den Berg, N. Vaessen, H.P. Endtz, T. Schulin, E.R. van der Vorm, and E.J. Kuijper, Evaluation of real-time PCR and conventional diagnostic methods for the detection of Clostridium difficile-associated diarrhoea in a prospective multicentre study. J Med Microbiol 56 (2007) 36-42.
- S. Bustin, Molecular medicine, gene-expression profiling and molecular diagnostics: putting the cart before the horse. Biomarkers in Medicine 2 (2008) 201-207.
- S.A. Bustin, Nucleic acid quantification and disease outcome prediction in colorectal cancer. Personalized Medicine 3 (2006) 207-216.
- S.A. Bustin, Real-time quantitative PCR-opportunities and pitfalls. EUROPEAN PHARMACEUTICAL REVIEW 4 (2008) 18-23.
- S.A. Bustin, V. Benes, T. Nolan, and M.W. Pfaffl, Quantitative real-time RT-PCR–a perspective. J Mol Endocrinol 34 (2005) 597-601.
- S.A. Bustin, and T. Nolan, Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J Biomol Tech 15 (2004) 155-66.
- S.A. Bustin, Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J. Mol. Endocrinol 29 (2002) 23-39.
- V. Uhlmann, C.M. Martin, O. Sheils, L. Pilkington, I. Silva, A. Killalea, S.B. Murch, J. Walker-Smith, M. Thomson, A.J. Wakefield, and J.J. O’Leary, Potential viral pathogenic mechanism for new variant inflammatory bowel disease. Mol Pathol 55 (2002) 84-90.
- M. Fitzpatrick, The end of the road for the campaign against MMR. British Journal of General Practice 57 (2007) 679.
- M.A. Afzal, L.C. Ozoemena, A. O’Hare, K.A. Kidger, M.L. Bentley, and P.D. Minor, Absence of detectable measles virus genome sequence in blood of autistic children who have had their MMR vaccination during the routine childhood immunization schedule of UK. J Med Virol 78 (2006) 623-30.
- Y. D’Souza, S. Dionne, E.G. Seidman, A. Bitton, and B.J. Ward, No evidence of persisting measles virus in the intestinal tissues of patients with inflammatory bowel disease. Gut 56 (2007) 886-8.
- Y. D’Souza, E. Fombonne, and B.J. Ward, No evidence of persisting measles virus in peripheral blood mononuclear cells from children with autism spectrum disorder. Pediatrics 118 (2006) 1664-75.
- M. Hornig, T. Briese, T. Buie, M.L. Bauman, G. Lauwers, U. Siemetzki, K. Hummel, P.A. Rota, W.J. Bellini, J.J. O’Leary, O. Sheils, E. Alden, L. Pickering, and W.I. Lipkin, Lack of association between measles virus vaccine and autism with enteropathy: a case-control study. PLoS ONE 3 (2008) e3140.
- T. Huang, H. Bohlenius, S. Eriksson, F. Parcy, and O. Nilsson, The mRNA of the Arabidopsis gene FT moves from leaf to shoot apex and induces flowering. Science 309 (2005) 1694-6.
- C.A. Perez-Novo, C. Claeys, F. Speleman, P. Van Cauwenberge, C. Bachert, and J. Vandesompele, Impact of RNA quality on reference gene expression stability. Biotechniques 39 (2005) 52, 54, 56.
- S. Fleige, and M.W. Pfaffl, RNA integrity and the effect on the real-time qRT-PCR performance. Mol Aspects Med 27 (2006) 126-39.
- S. Fleige, V. Walf, S. Huch, C. Prgomet, J. Sehm, and M.W. Pfaffl, Comparison of relative mRNA quantification models and the impact of RNA integrity in quantitative real-time RT-PCR. Biotechnol Lett 28 (2006) 1601-13.
- T. Nolan, R.E. Hands, B.W. Ogunkolade, and S.A. Bustin, SPUD: a qPCR assay for the detection of inhibitors in nucleic acid preparations. Anal Biochem 351 (2006) 308-310.
- T. Nolan, R.E. Hands, and S.A. Bustin, Quantification of mRNA using real-time RT-PCR. Nature Protocols 1 (2006) 1559-1582.
- C. Tricarico, P. Pinzani, S. Bianchi, M. Paglierani, V. Distante, M. Pazzagli, S.A. Bustin, and C. Orlando, Quantitative real-time reverse transcription polymerase chain reaction: normalization to rRNA or single housekeeping genes is inappropriate for human tissue biopsies. Anal. Biochem. 309 (2002) 293-300.
- J. Huggett, K. Dheda, S. Bustin, and A. Zumla, Real-time RT-PCR normalisation; strategies and considerations. Genes Immun 6 (2005) 279-84.
- K. Dheda, J.F. Huggett, J.S. Chang, L.U. Kim, S.A. Bustin, M.A. Johnson, G.A. Rook, and A. Zumla, The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal Biochem 344 (2005) 141-3.
- K. Dheda, J.F. Huggett, S.A. Bustin, M.A. Johnson, G. Rook, and A. Zumla, Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques 37 (2004) 112-119.
- J. Vandesompele, K. De Preter, F. Pattyn, B. Poppe, N. Van Roy, A. De Paepe, and F. Speleman, Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3 (2002) 0034.1-0034.11.
- M.W. Pfaffl, A. Tichopad, C. Prgomet, and T.P. Neuvians, Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol Lett 26 (2004) 509-15.
- C.L. Andersen, J.L. Jensen, and T.F. Orntoft, Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64 (2004) 5245-50.
- S.A. Bustin, J. Garson, J. Hellemans, J. Hugett, M. Kubista, R. Mueller, T. Nolan, M. Pfaffl, G.L. Shipley, J. Vandesompele, and C.T. Wittwer, The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clinical Chemistry (2009) (submitted).
- A. Brazma, P. Hingamp, J. Quackenbush, G. Sherlock, P. Spellman, C. Stoeckert, J. Aach, W. Ansorge, C.A. Ball, H.C. Causton, T. Gaasterland, P. Glenisson, F.C. Holstege, I.F. Kim, V. Markowitz, J.C. Matese, H. Parkinson, A. Robinson, U. Sarkans, S. Schulze-Kremer, J. Stewart, R. Taylor, J. Vilo, and M. Vingron, Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat.Genet. 29 (2001) 365-371.
- C.F. Taylor, N.W. Paton, K.S. Lilley, P.A. Binz, R.K. Julian, Jr., A.R. Jones, W. Zhu, R. Apweiler, R. Aebersold, E.W. Deutsch, M.J. Dunn, A.J. Heck, A. Leitner, M. Macht, M. Mann, L. Martens, T.A. Neubert, S.D. Patterson, P. Ping, S.L. Seymour, P. Souda, A. Tsugita, J. Vandekerckhove, T.M. Vondriska, J.P. Whitelegge, M.R. Wilkins, I. Xenarios, J.R. Yates, 3rd, and H. Hermjakob, The minimum information about a proteomics experiment (MIAPE). Nature Biotechnology 25 (2007) 887-93.
- D. Field, G. Garrity, T. Gray, N. Morrison, J. Selengut, P. Sterk, T. Tatusova, N. Thomson, M.J. Allen, S.V. Angiuoli, M. Ashburner, N. Axelrod, S. Baldauf, S. Ballard, J. Boore, G. Cochrane, J. Cole, P. Dawyndt, P. De Vos, C. DePamphilis, R. Edwards, N. Faruque, R. Feldman, J. Gilbert, P. Gilna, F.O. Glockner, P. Goldstein, R. Guralnick, D. Haft, D. Hancock, H. Hermjakob, C. Hertz-Fowler, P. Hugenholtz, I. Joint, L. Kagan, M. Kane, J. Kennedy, G. Kowalchuk, R. Kottmann, E. Kolker, S. Kravitz, N. Kyrpides, J. Leebens-Mack, S.E. Lewis, K. Li, A.L. Lister, P. Lord, N. Maltsev, V. Markowitz, J. Martiny, B. Methe, I. Mizrachi, R. Moxon, K. Nelson, J. Parkhill, L. Proctor, O. White, S.A. Sansone, A. Spiers, R. Stevens, P. Swift, C. Taylor, Y. Tateno, A. Tett, S. Turner, D. Ussery, B. Vaughan, N. Ward, T. Whetzel, I. San Gil, G. Wilson, and A. Wipat, The minimum information about a genome sequence (MIGS) specification. Nature Biotechnology 26 (2008) 541-7.
- C.J. Echeverri, P.A. Beachy, B. Baum, M. Boutros, F. Buchholz, S.K. Chanda, J. Downward, J. Ellenberg, A.G. Fraser, N. Hacohen, W.C. Hahn, A.L. Jackson, A. Kiger, P.S. Linsley, L. Lum, Y. Ma, B. Mathey-Prevot, D.E. Root, D.M. Sabatini, J. Taipale, N. Perrimon, and R. Bernards, Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat Methods 3 (2006) 777-9.
- S.A. Haney, Increasing the robustness and validity of RNAi screens. Pharmacogenomics 8 (2007) 1037-49.
- S.A. Sansone, T. Fan, R. Goodacre, J.L. Griffin, N.W. Hardy, R. Kaddurah-Daouk, B.S. Kristal, J. Lindon, P. Mendes, N. Morrison, B. Nikolau, D. Robertson, L.W. Sumner, C. Taylor, M. van der Werf, B. van Ommen, and O. Fiehn, The metabolomics standards initiative. Nature Biotechnology 25 (2007) 846-8.
- C.F. Taylor, D. Field, S.A. Sansone, J. Aerts, R. Apweiler, M. Ashburner, C.A. Ball, P.A. Binz, M. Bogue, T. Booth, A. Brazma, R.R. Brinkman, A. Michael Clark, E.W. Deutsch, O. Fiehn, J. Fostel, P. Ghazal, F. Gibson, T. Gray, G. Grimes, J.M. Hancock, N.W. Hardy, H. Hermjakob, R.K. Julian, Jr., M. Kane, C. Kettner, C. Kinsinger, E. Kolker, M. Kuiper, N.L. Novere, J. Leebens-Mack, S.E. Lewis, P. Lord, A.M. Mallon, N. Marthandan, H. Masuya, R. McNally, A. Mehrle, N. Morrison, S. Orchard, J. Quackenbush, J.M. Reecy, D.G. Robertson, P. Rocca-Serra, H. Rodriguez, H. Rosenfelder, J. Santoyo-Lopez, R.H. Scheuermann, D. Schober, B. Smith, J. Snape, C.J. Stoeckert, Jr., K. Tipton, P. Sterk, A. Untergasser, J. Vandesompele, and S. Wiemann, Promoting coherent minimum reporting guidelines for biological and biomedical investigations: the MIBBI project. Nature Biotechnology 26 (2008) 889-96.
- J. Hellemans, G. Mortier, A. De Paepe, F. Speleman, and J. Vandesompele, qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8 (2007) R19.
- S. Lefever, J. Hellemans, F. Pattyn, D.R. Przybylski, C. Taylor, R. Geurts, A.V. Untergasser, and J. Vandesompele, RDML: structured language and reporting guidelines for real-time quantitative PCR data. Nucleic Acids Res (submitted) (2009).
- H.D. VanGuilder, K.E. Vrana, and W.M. Freeman, Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques 44 (2008) 619-26.
- M.W. Pfaffl, I.G. Lange, A. Daxenberger, and H.H. Meyer, Tissue-specific expression pattern of estrogen receptors (ER): quantification of ER alpha and ER beta mRNA with real-time RT-PCR. 109 (2001) 345-355.
- M.W. Pfaffl, A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29 (2001) E45.
- S.N. Peirson, J.N. Butler, and R.G. Foster, Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. Nucleic Acids Res 31 (2003) e73.
- C. Ramakers, J.M. Ruijter, R.H. Deprez, and A.F. Moorman, Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339 (2003) 62-6.
- W. Liu, and D.A. Saint, Validation of a quantitative method for real time PCR kinetics. Biochem.Biophys.Res.Commun. 294 (2002) 347-353.
- R.G. Rutledge, Sigmoidal curve-fitting redefines quantitative real-time PCR with the prospective of developing automated high-throughput applications. Nucleic Acids Res 32 (2004) e178.
- R.G. Rutledge, and C. Cote, Mathematics of quantitative kinetic PCR and the application of standard curves. Nucleic Acids Res 31 (2003) e93.
- R.G. Rutledge, and D. Stewart, A kinetic-based sigmoidal model for the polymerase chain reaction and its application to high-capacity absolute quantitative real-time PCR. BMC Biotechnol 8 (2008) 47.
- A. Tichopad, M. Dilger, G. Schwarz, and M.W. Pfaffl, Standardized determination of real-time PCR efficiency from a single reaction set-up. Nucleic Acids Res 31 (2003) e122.
- S. Zhao, and R.D. Fernald, Comprehensive algorithm for quantitative real-time polymerase chain reaction. Journal of Computational Biology 12 (2005) 1047-64.
- C. Ritz, and A.N. Spiess, qpcR: an R package for sigmoidal model selection in quantitative real-time polymerase chain reaction analysis. Bioinformatics 24 (2008) 1549-51.
- A. Batsch, A. Noetel, C. Fork, A. Urban, D. Lazic, T. Lucas, J. Pietsch, A. Lazar, E. Schomig, and D. Grundemann, Simultaneous fitting of real-time PCR data with efficiency of amplification modeled as Gaussian function of target fluorescence. BMC Bioinformatics 9 (2008) 95.
- D.V. Rebrikov, and D.Y. Trofimov, Real-time PCR: A review of approaches to data analysis Applied Biochemistry and Microbiology 42 (2006) 455-463.
- J.M. Gallup, and M.R. Ackermann, Addressing fluorogenic real-time qPCR inhibition using the novel custom Excel file system ‘FocusField2-6GallupqPCRSet-upTool-001’ to attain consistently high fidelity qPCR reactions. Biol Proced Online 8 (2006) 87-152.
- J.M. Gallup, and M.R. Ackermann, The ‘PREXCEL-Q Method’ for qPCR International Journal of Biomedical Science 4 (2008) 100-120.
- P.Y. Muller, H. Janovjak, A.R. Miserez, and Z. Dobbie, Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques 32 (2002) 1372-1379.
- N. Jin, K. He, and L. Liu, qPCR-DAMS: a database tool to analyze, manage, and store both relative and absolute quantitative real-time PCR data. Physiol Genomics 25 (2006) 525-7.




