Finding novel targets for anticancer target discovery
Posted: 9 May 2010 |
Advances in our understanding of the molecular basis of cancer together with novel approaches to interfere with signal transduction pathways have opened new horizons for anticancer target discovery. In particular, the image based large scale analysis of cellular phenotypes that arise from genetic or chemical perturbations paved the way for the identification and validation of disease relevant molecular targets independent of preconceived notions of mechanistic relationships.


Advances in our understanding of the molecular basis of cancer together with novel approaches to interfere with signal transduction pathways have opened new horizons for anticancer target discovery. In particular, the image based large scale analysis of cellular phenotypes that arise from genetic or chemical perturbations paved the way for the identification and validation of disease relevant molecular targets independent of preconceived notions of mechanistic relationships.
Today, more than 80 years after the discovery of penicillin which revolutionised the therapy of infectious diseases, we continue pursuing a similar breakthrough for the treatment of cancer. Unfortunately, most cancers do not possess molecular targets comparable to the bacterial cell wall (the target of penicillin), which is essential and specific for the bacteria1. The success of conventional cancer therapy is limited as a result of the lack of selectivity between cancer cells and normal cells.
From conventional chemotherapy to molecularly targeted therapies
The development of the first cancer chemo – therapy dates back to the early 1940s when the anticancer activity of the nitrogen mustard-based war gas was discovered. Nitrogen mustards are DNA alkylating agents that interfere with DNA replication. Like the DNA alkylating agents, the most frequently used chemotherapy drugs have been identified empirically without pre-existing knowledge of the molecular mechanism of action2. Despite the groundbreaking progress in the understanding of cancer, including the characterisation of cancer viruses of animals in the 1960s and 1970s, the discovery of oncogenes and tumour suppressor genes in the 1970s and 1980s and the more recent mapping of signalling networks associated with the hallmarks of cancer3, the treatment options for cancer haven’t changed much in past decades. Certain malignancies including testicular cancer, gestational choriocarcinoma, Hodgkin’s disease and high-grade lymphomas have shown to be highly sensitive to conventional chemotherapy. Optimisation and combination of conventional cytotoxic chemotherapy have produced higher cure rates for many cancers. By contrast, curative treatment remains elusive for the majority of patients with metastatic tumours4. In the absence of a specific mechanistic understanding, particularly with regard to the molecular target, it is difficult to learn from the successes and failures and understand why different tumour types have different susceptibilities2. However, the cancer-research community acquired a critical mass of significant knowledge that allowed for the transition of modern anticancer drug discovery from cytotoxic towards targeted therapies. Targeted therapeutic agents, including small molecules or monoclonal antibodies, interfere with a discreet molecular target in order to have desired therapeutic effects and have the potential to be personalised for each patient.
Gleevec (Imatinib), a kinase inhibitor that was introduced in 2001 for the treatment of chronic myelogenous leukaemia (CML), emerged as a paradigm for molecularly targeted therapies. Other examples of the successful application of targeted therapies include the genetically engineered antibodies trastuzumab and rituximab, which are available for the treatment of metastatic breast cancer and non-Hodgkin’s Lymphoma, respectively. Gleevec was identified as an inhibitor of the platelet-derived growth factor (PDGF) receptor kinase, but was soon recognised to inhibit c-Abl, a kinase which is frequently altered in chronic myelogenous leukaemia (CML). This disease is caused by an acquired translocation between chromosome 9 and chromosome 22 that produces fusion of the Bcr and Abl proteins. BCR-ABL fusion results in a constitutively active tyrosine kinase enzyme that initiates CLM (Figure 1). The spectacular success of Gleevec has proven that the concept of targeting specific molecular genetic events in cancer can result in highly efficient anticancer therapies. Nevertheless, as CML is a genetically simple neoplasm caused by a single aberrant protein, there is still substantial debate about whether the Gleevec paradigm can be translated to other cancers which are caused by a multitude of complex interacting genetic and environmental factors5. In order to tackle the complex derangement present in the majority of cancers, the treatment has to be intimately linked to the causal changes for each neoplasm. Hence, the selection of appropriate combinations of molecular targets specific for different cancers will be essential to develop future cancer therapeutics with new modes of action that selectively kill the cancer cells.
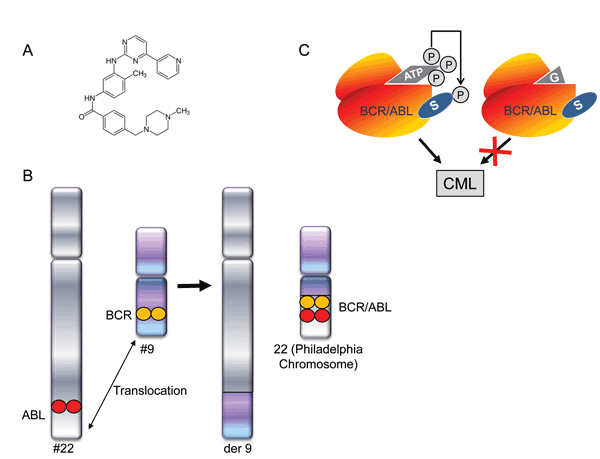

Figure 1
Target discovery in the postgenomic era
The identification and early validation of disease- relevant molecular targets is an essential first step in the drug discovery pipeline. While the sequencing of the human genome and the emerging data on cancer genomes represent substantial progress in identifying the genetic alterations in human cancer, considerable work remains in gaining an understanding of how certain gene products drive tumorigenesis. The complexity of the signalling networks in which these proteins operate has ushered in the need for unbiased target discovery strategies independent of preconceived notions of mechanistic relationships. There are a significant number of emerging technologies attempting to deal with the complexity of intact cells and shed light on the signal transduction mechanisms underlying tumorigenesis and cancer progression. This review will focus on the use of cell-based high content imaging of cellular phenotypes that arise from genetic or chemical perturbations.
High content screening as an essential tool for target discovery
High content screening was introduced to combine the efficiency of high throughput techniques with the ability of cellular imaging to collect quantitative data from complex biological systems6. The ability to measure at the single cell level and to select a cell or a subpopulation of cells of interest is one of the most conspicuous features of HCS. At the same time, the level of complexity which can be addressed with HCS is outstanding among the various large scale approaches. The complexity of the cellular phenotype is reflected in the wealth of information present in most HCS datasets. Current available algorithms can be readily applied for the automated extraction of multi-dimensional information from cellular images suitable to characterise many different phenotypical events which fall into four categories, namely fluorescence intensity changes, fluorescence distribution, morphology and cell movement. In other words, HCS is capable of cracking open the black box of the cell. Diverse responses to perturbing agents such as small molecule compounds or RNAi can be monitored simultaneously, a process that was coined as multiplexing. The acquired quantitative and qualitative information such as number, intensity, size, morphology, texture and spatial distribution of the objects can be computed flexibly and the archived data can be reanalysed with different variables and combinations of parameters according the needs of the particular drug discovery or basic research program.
HCS has been widely used to support interference strategies for target discovery and validation by using pharmaceutical inhibitors or chemical agents. Aside from its use as starting points for drug discovery, small organic molecules have proven to be invaluable tools to perturb biology and investigate biological function in basic research7. This approach, widely referred to as chemical genetics, was largely motivated by the need to determine the function of the large number of genes identified by the human genome sequencing project and the unique role of small molecules as physiological regulators of protein function. The combination of chemical genetics and HCS has turned out to be a powerful tool for uncovering protein functions and dissecting signalling pathways. RNAi technology is another method to probe gene function systematically, which has become compatible with mammalian cell culture and has ushered in the need for cell based technologies suitable for high throughput analysis in the academic research environment. RNAi-mediated gene silencing effect depends on several parameters including transfection efficiency, the intrinsic expression rate of the mRNA and the turnover rate of the protein. Furthermore, many RNAi’s produce only partial knockdowns. Therefore, RNAi-based HCS experiments tend to be noisier compared with those using pharmacological inhibition and considerable effort has to be expended to confirm specificity of the RNAi knockdown effects. Several studies report successful genome-wide or subgenome-wide high content screens combined with thorough follow up analysis ensuring target specificity8-10.
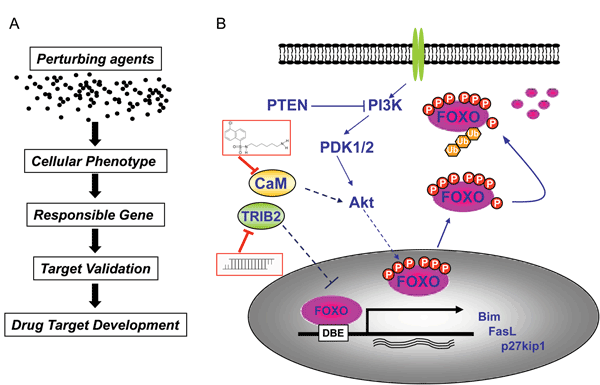

Figure 2
Finding novel molecular targets associated with the PI3K/Akt/FOXO signalling axis
Our laboratory is particularly interested in the regulation of FOXO proteins. FOXO transcription factors are bona fide tumour suppressors that are inactivated in the majority of human cancers, owing to the overactivation of the phosphoinositide 3-kinase (PI3K)/Akt pathway11,12. Reactivation of FOXO based on its tumour suppressor properties is considered a very attractive anti-cancer strategy. Contrary to other tumour suppressors such as p53 or PTEN, the functions of which are abrogated by genetic or epigenetic changes, inactivation of FOXOs occurs mostly because of the overactivation of their inhibitory inputs11,12. This offers a wide range of possibilities for restoring FOXO activity with small-molecule inhibitors targeting up-regulated FOXO repressors. In order to identify possible targets for the restoration of FOXO functions, we employed cell-based screening systems to systematically interrogate the signalling network that controls the activity of FOXO proteins.
Chemical genetic approach to target discovery
We applied a chemical biology approach to study the mechanisms that influence the intracellular localisation of the FOXO family member FOXO3a. We established a high-throughput cellular-imaging assay that monitors the nuclear–cytoplasmic translocation of a GFP–FOXO3a fusion protein in tumour cells. We identified 17 compounds from a panel of 73 low-molecular weight compounds with known biological activity capable of inducing the nuclear accumulation of GFP–FOXO. As expected, these compounds included chemicals known to interfere with components of the PI3K/Akt signalling pathway, as well as with nuclear export. Interestingly, the calmodulin antagonists W-7, W-13 and calmidazolium chloride (CDZ) also produced a positive result in the HCS assay. We were able to show that the molecular mechanisms by which the CaM antagonists exert their effect on FOXO trans – location were mediated by the CaM-mediated decrease of Akt activity13. The identification of CaM as a molecular target that could be exploited pharmacologically to reactivate FOXO activity in human tumours illustrates the power of chemical genetics combined with image-based cellular screening to analyse signalling pathways.
Finding novel targets using HCS-based RNAi screening
The advent of RNAi-based functional genomics in mammals enabled the conduction of unbiased screens in human cell systems. In order to identify endogenous suppressors of FOXO, which a priori have the potential to be targeted to restore FOXO functions, we performed a systematic large-scale loss of function screening factors. Based on two independent cell-based systems that were shown to effectively report FOXO-dependent transcriptional activity and subcellular localisation of the GFP–FOXO fusion protein, we screened a vector-based RNAi expression library that directs the synthesis of shRNAs targeting for suppression of 7914 different human genes10. Among the genes identified and validated as negative FOXO regulators, we found the pseudokinase TRIB2, the human homolog of the Drosophila gene tribbles. We found TRIB2 to be highly expressed specifically in melanoma malignancies. In vitro and in vivo results reported here indicate that TRIB2 can facilitate the growth and survival of melanomas by downregulating FOXO activity. Melanoma lesions are the most aggressive form of skin cancer, resistant to all standard anticancer therapies14. Further investigation into the role of TRIB2 in the pathogenesis of melanoma lesions may provide new therapeutic insights and lead to the development of TRIB2 as a therapeutic target for melanoma.
Conclusions
A new arsenal of therapeutic options selectively targeting cancers without toxicity to normal tissues is urgently needed. The successful treatment of chronic myeloid leukaemia with Gleevec has provided the proof of principle for the concept of targeted anti-cancer therapy. The identification of molecular targets intimately linked to the causal changes in the major human malignancies will be essential to develop new strategies for the targeted treatment of cancer. The application of RNA interference and chemical genetics combined with image-based cellular screening has proven a powerful tool for the unbiased identification of disease relevant molecular targets. Acknowledgements This work was supported by a grant from the Spanish MEC (project BIO2006-02432).
References
- M. V. Blagosklonny, Drug Discov Today 8, 1104 (Dec 15, 2003)
- J. B. Gibbs, Science 287, 1969 (Mar 17, 2000)
- D. Hanahan, R. A. Weinberg, Cell 100, 57 (Jan 7, 2000)
- P. Savage, J. Stebbing, M. Bower, T. Crook, Nat Clin Pract Oncol 6, 43 (Jan, 2009)
- W. G. Kaelin, Jr., Sci STKE 2004, pe12 (Mar 23, 2004)
- V. C. Abraham, D. L. Taylor, J. R. Haskins, Trends Biotechnol 22, 15 (Jan, 2004)
- B. R. Stockwell, Nature 432, 846 (Dec 16, 2004)
- J. Moffat et al., Cell 124, 1283 (Mar 24, 2006)
- J. Moffat, D. M. Sabatini, Nat Rev Mol Cell Biol 7, 177 (Mar, 2006)
- F. Zanella et al., Oncogene, (Mar 8, 2010)
- T. B. Dansen, B. M. Burgering, Trends Cell Biol 18, 421 (Sep, 2008)
- F. Zanella, W. Link, A. Carnero, Curr Cancer Drug Targets, (Jan 1, 2010)
- F. Zanella, A. Rosado, B. Garcia, A. Carnero, W. Link, Chembiochem 9, 2229 (Sep 22, 2008)
- M. S. Soengas, S. W. Lowe, Oncogene 22, 3138 (May 19, 2003)
About the author
Dr. Wolfgang Link currently holds a position at the Department of Experimental Therapeutics at the Spanish National Cancer Centre (CNIO) in Madrid (Spain) where he is responsible for the image based translational research aiming to discover anti-cancer therapies. Dr. Wolfgang Link is credited with two patents and 27 publications in leading academic journals. His research led to the discovery of essential genes in synaptic plasticity and pain control. Upon completion of his postdoctoral training at the CNB (Madrid, Spain), he moved into the biotechnology industry in Munich (Germany). Wolfgang joined the CNIO in 2001.




