Proteomics – the frontiers and beyond
Posted: 21 September 2007 | | No comments yet
Within a decade proteomics has evolved from a fledgling discipline reserved for specialised laboratories, to a firm fixture in our standard omics arsenal used routinely by the research community. This stunning progress is due to many factors; the finishing of the genome projects provided major intellectual motivation and the development of better and much easier to use mass spectrometry (MS) instruments were technological drivers. What is even more remarkable is that this progress has been made despite leaving some main issues in proteomics unsolved. Besides these known boundaries, it also has revealed new frontiers and new interesting glimpses into the world beyond. This essay discusses some selected issues, but cannot necessarily be exhaustive or free of personal opinion.
Within a decade proteomics has evolved from a fledgling discipline reserved for specialised laboratories, to a firm fixture in our standard omics arsenal used routinely by the research community. This stunning progress is due to many factors; the finishing of the genome projects provided major intellectual motivation and the development of better and much easier to use mass spectrometry (MS) instruments were technological drivers. What is even more remarkable is that this progress has been made despite leaving some main issues in proteomics unsolved. Besides these known boundaries, it also has revealed new frontiers and new interesting glimpses into the world beyond. This essay discusses some selected issues, but cannot necessarily be exhaustive or free of personal opinion.
Within a decade proteomics has evolved from a fledgling discipline reserved for specialised laboratories, to a firm fixture in our standard omics arsenal used routinely by the research community. This stunning progress is due to many factors; the finishing of the genome projects provided major intellectual motivation and the development of better and much easier to use mass spectrometry (MS) instruments were technological drivers. What is even more remarkable is that this progress has been made despite leaving some main issues in proteomics unsolved. Besides these known boundaries, it also has revealed new frontiers and new interesting glimpses into the world beyond. This essay discusses some selected issues, but cannot necessarily be exhaustive or free of personal opinion.
What is the relationship between genome and proteome?
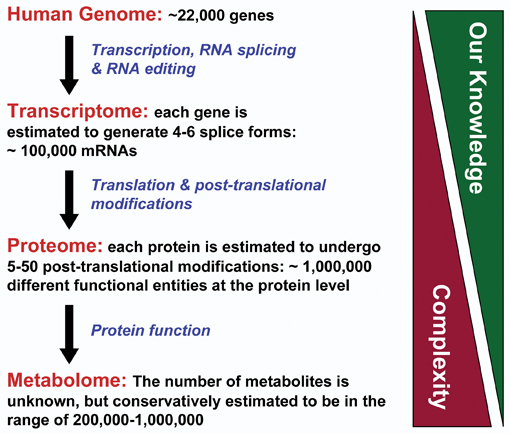
The completion of the genome projects has somewhat ironically clarified that the answers to many of the big questions in biology and medicine lie in the proteins rather than the genes. Take the human genome for example; with a mere 21,000 odd genes, less or similar to some common plants, we can produce 100,000 or more functionally different proteins1. In addition, many proteins are multifunctional and moonlight in many different jobs within the cell. Therefore, we need to look closely at the proteins, and have made enormous strides towards that in a short period of time. Here, the availability of reliable genome sequences has enabled proteomics by providing the databases from which MS experiments can extract the information for routine protein identification. On the other hand, proteomics feeds back into genomics by helping to delineate the boundaries of genes, identifying splice forms and understanding gene regulation and genomic maintenance. Thus, genomics and proteomics seem close companions.
Yet there is huge rivalry when the question comes to where the regulation takes place. It is proteins that transcribe the genes, and it is proteins which regulate the proteins that transcribe genes. However, the discovery of micro (mi)RNAs and small inhibitory (si)RNAs has revealed a whole new world of genes regulating genes2. While admittedly they still use proteins to do their job, all of the regulatory instructions are provided by the miRNAs and siRNAs. Also, when analysing gene array data, an emerging picture is that transcription factors regulate other transcription factors, which regulate yet other transcription factors. In the very few instances where transcriptomic and proteomic datasets have been put together, there is a tendency for rather good correlation in single cell organisms and poor correlation in multicellular and complex organisms. Changing the food source will hugely change the transcriptional and proteomic profile of a single celled organism, because the transcriptional units are organised to express whole genetic programmes. Contrastingly, multicellular organisms’ transcription and protein function seems to be rather independently regulated. Thus, in terms of regulation, transcriptome and proteome currently present themselves as two largely de-coupled networks. From an engineering perspective this concept is appealing as its robustness helps to prevent a failure in one network from automatically spreading into the other. However, from a biological point of view this concept is rather hard to believe. It is difficult to explain how biological evolution, responding to the same environmental constraints would have split the co-regulation and come up with independent designs for two different networks and yet make them work together seamlessly. It appears we still have a lot to learn.
It is not clear at present what the best approach will be to crack the riddle of the relationship between genome and proteome. One promising idea is to apply proteomics to genetic model organisms such as; yeast, the fruit fly Drosophila melanogaster and the worm C. elegans. These organisms have well explored genetics and are amenable to genetic manipulation, with thousands of characterised mutant phenotypes available. Their main advantage is that genetic mutations can be correlated with changes in the proteome and a biological phenotype. Huge explanatory power is derived when proteome analysis becomes the intellectual glue between genotype and phenotype. The establishment of large scale protein interaction networks in these organisms will hopefully be followed by a systematic analysis of mutant phenotypes on proteome and transcriptome levels. This will be a real opportunity to filter out the connections that generate biological function.
The science of change
The above paragraph implies that we should make lots of comparisons. Normally in this type of analyses we compare steady states, but is that enough? Maybe this is why genomic and proteomic regulation appears de-coupled? As an example consider that there is a time delay between transcription of a gene and translation into protein and that this delay can vary greatly between different genes. Therefore, by looking at steady states we face completely unsynchronised scales that will not readily reveal the connections between transcription and translation. Perhaps we should look at rates of changes; this could also at least in part explain how genomic and proteomic regulatory networks can continuously communicate without the risk of a melt down due to fatal error propagation, through hardwired communication channels that are always open. Soft-wiring communication channels by synchronising them on concurrent dynamic events would be a powerful filter for ensuring that communication and signal propagation is appropriate to and licensed by the current state of a cell. This would mean that erroneous communications would be cut out.
In order to test this hypothesis we will need to be able to measure the rates of change. There are a number of quantitative proteomics methods that can determine rates of changes in protein expression, modifications, synthesis and decay. However, these measurements suffer from the ubiquitous limitation of proteomics. For example; we can usually only survey a small fraction of the proteome, but we can do this in great detail and even determine post-translational modifications (PTMs). In contrast, we can routinely assay on a global scale changes in mRNA levels. However, here we lack detail, as we cannot establish whether changes in mRNA abundance are due to differences in transcriptional rates or stability. So we generate data that are quite different in their nature, structure and quality, and it will be a formidable challenge to put them together in an intelligible way. This will require new methods in bioinformatics and especially new methods for meta-analysis and analysis of dynamic systems that will borrow concepts from engineering, mathematics and physics. In fact, the biggest deficit is probably the lack of a clear conceptual framework. The blooming field of systems biology will no doubt contribute its efforts to this and it is no surprise that proteomics has provided a major impetus for systems biology. Thus, we can look ahead to a stimulating relationship.
Disentangling complexity: divide and conquer!
As previously discussed the biggest probable limitation of proteomics is that in routine experiments we only can analyse a small fraction of the proteome, which is usually the most abundant protein. This is due to the enormous complexity and dynamic range of the proteome. This issue has proven difficult to address, and as far as we can tell there will be more than one answer. As none is perfect, a combination of orthogonal methods is likely to provide the long term solutions. The current, most popular and very logical answer is to chop the proteome in more palatable fractions by improving pre-separation methods before MS analysis. However, most of the separation methods are biased towards certain physico-chemical properties of proteins, and virtually all carry a severe penalty for speed.
The former drawback can actually be turned into an advantage as demonstrated by the Vandekerckhove and Gevaert groups who developed COFRADIC3. In this method peptides are separated by LC, but then the peptide fractions are subjected to chemical or biochemical treatment that changes the chromatographic behaviour of successfully modified peptides. These modified peptides can now be isolated by re-chromatography of modified fractions on the same columns for identification by MS. The power of this approach lies in its versatility and its ability to efficiently reduce complexity. There is virtually no limit to employing modification strategies that target different PTMs or selected amino acids including those which can provide positional information, for example selection for N-terminal peptides. Thus, the method is enormously useful. High efficiency separation can be achieved, but experiments can take days or even weeks. Thus, there is still a need for faster, efficient and MS compatible separation methods.
One approach that fulfils these criteria is actually MS itself, as high resolution MS is a very powerful separation device. Hybrid instruments that combine two or even three MS methods permit to isolate, resolve and analyse hundreds of thousands of peptide species. Although this cannot be explored in great detail, it is important to be aware that it is an active area of development where superior solutions are constantly becoming available. A principal drawback of these methods is the MS bias towards the small mass range. Thus, the conversion of proteins to peptides by proteolytic digestion and the subsequent MS compatible fractionation of peptides is a necessary part of most of these methods. An epitome of this approach is the shotgun sequencing of multi-dimensionally separated peptide digests, which was originally developed by the Yates laboratory as a technology called MudPIT4, but now comes in several different forms. Here, a proteolytic digest of proteins is prepared and peptides are separated by an ion exchanger followed by a reverse phase column. The fractions are analysed by MS. This approach is very useful for the analysis of medium complexity samples (a few hundred to a few thousand proteins), where it gives nearly exhaustive identifications. However, the digestion step increases complexity and experiments are difficult to compare when the complexity of the sample exceeds the separation capacity. In addition, the digestion largely destroys information on protein isoforms and the patterns of PTMs on the individual protein level. In principle, this could be circumvented by the two-dimensional separation and MS analysis of whole proteins instead of peptides. However, the lack of suitable separation media and technical constraints of ‘top-down’ MS of intact proteins have hampered this approach.
New developments are now on the horizon; the IRColl RASOR (Interdisciplinary Research Collaboration for Radical Solutions for Researching the proteome), consortium between the Universities of Glasgow, Edinburgh and Dundee, Scotland and UK, is dedicated to developing innovative technologies that will be generically useful, but are beyond the reach of current technologies. Combining new chromatographic separation media (from Dionex) with high resolution Fourier Transform (FT), (MS ,from Bruker Daltonics and, RASOR) has developed a technology platform that per experiment could separate and identify ca. 20% of the predicted protein complement of bacteria. This is comparable to peptide based shotgun approaches and will further improve with better software for top-down proteomics. Importantly, the ability to analyse intact proteins gives the unique opportunity to study PTMs on the level of individual protein species. What remains a major challenge is the inherent MS bias towards smaller protein masses. Amongst possible solutions are improvements in the MS ionisation techniques and more efficient and sensitive methods for protein fragmentation that will extend the MS mass range. Further, the combination of methods that can separate individual protein species of higher mass, but then perform MS analysis on the peptide level (such as 2D gel based proteomics), could be very useful.
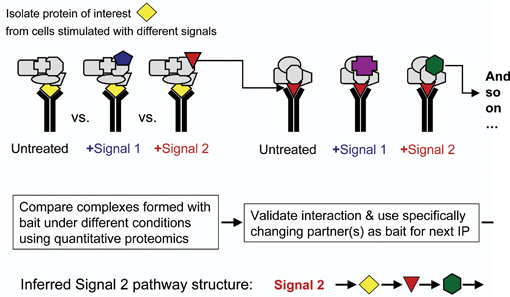
In the meantime, a powerful approach is interaction proteomics. The advantages are twofold. First, complexity is reduced by several orders of magnitude when we selectively look at the proteins that associate with a given bait protein. Second, we can glean some functional information when we monitor the changes in protein interactions using one of the quantitative methods. Using cross-comparisons makes this approach very powerful indeed. It lends itself to track the flow of a signal through a network by simply monitoring the changes in protein interactions. Conceptually this is a biochemical equivalent of genetic epistasis experiments, and equally powerful. Using this approach we have mapped an apoptotic pathway over six steps from the cell membrane to nuclear transcription5. Thus, the ‘interaction proteomics approach’ is and will be prolific in the future. An extension to the MS based analytic end is now afforded by protein interaction array- based proteomics. This technology has jumped the initial technical hurdles and is now quickly becoming established. A potential advantage is the ease of use and no requirement for expensive MS equipment. In very limited comparisons we have seen little overlap between MS and array based interactome studies. Thus, lesson is that no method is perfect, and that like a musical orchestra we will need to use different instruments to play a harmonic piece.
Small is beautiful!
A typical proteomics experiment requires between millions and billions of cells. Ideally we would like to analyse what is going on in a few or even a single cell. There are many good reasons to justify this quest; first is the availability of only a small amount of sample, for example, from clinical biopsy material. Second, is the issue that measurements at the population level do not necessarily reflect the behaviour of the individual cells. This becomes very relevant for stem cell research where daughter cells may adopt different fates and where such decisions are often governed by growth factor gradients. For instance, when we analyse a population of cells, a graded response to an increasing stimulus can arise by each cell responding in a graded fashion, or by a change in the fraction of cells responding in an ‘all or nothing’ manner. This only can be decided by single cell studies. Are they in reach?
The sensitivity of MS has been steadily increased over the years and further improvements are possible, for example, enhancing ionisation efficiencies and detector speed. Another important area is the optimisation of sample preparation and separation; as with small samples, huge losses occur due to protein adsorption during sample handling. An appealing solution is to minimise sample loss by miniaturising the handling and integrating the lysis and separation step into one platform. Such lab-on-a-chip technology is a flourishing area of development, where we can expect elegant solutions. However, high resolution separation and the desirable direct online link to MS analysis remain challenges. There are numerous efforts around the world, including the RASOR consortium, which focus on; miniaturising affinity capture approaches as a separation medium, and controlled desolvation as means for delivering minute amount of samples with high efficiency into the MS. Ideally, we are aiming for the analysis of single cells that are trapped using optical tweezers, lysed in situ with online separation of their protein complement and subsequent feed into the MS. That dream may not be far off as most of the individual components already exist, but still need to be integrated into a single micro-fluidic device. This still is a formidable task, but the silver lining is on the horizon.
Another very attractive approach is imaging. We have been looking down microscopes for centuries now, and still there are significant technical improvements and new discoveries. The main limitation of microscopy only delivering purely descriptive data is now being overcome by the development of methods that inject function into imaging. Methods based on Fluorescence Resonance Energy Transfer (FRET) or Fluorescence Cross Correlation Spectroscopy (FCCS) can detect and quantify protein interactions in single cells at single molecule sensitivity. When combined with “smart” probes such as fluorescent phosphatase or kinase substrates, biochemical reactions can be monitored in single living cells with spatial resolution. This is a quantum leap forward. The flipside of the coin is that these methods require expensive equipment, are complicated and error prone and hence still the domain of a few specialist laboratories. This was the situation with MS based proteomics 10 years ago, and I think we can look forward to an era of imaging based proteomics. An interesting variation on this theme is the combination of MS and imaging. The use of MS to image the mass distribution of peptides in tissues was pioneered by Richard Caprioli’s group6. It permits the correlation of anatomical features with the distribution of specific marker peptides and also the monitoring of drug distribution and metabolism in tissues. The functional aspect of this technology of course hinges on the identification of the marker peptides. This is difficult, but is now being solved using MS/MS approaches. The spatial resolution is currently rather poor (still less than the size of a single cell), but this may be solved by improving laser technology and by combing MS imaging with laser micro-dissection which permits the physical isolation of single cells or cell units from tissues.
Outlook
Where are we going from here? The next big challenge will be to produce the functional data that can feed into systems biology. Here, the circle closes as proteomics was one of the trail blazers for systems biology. There are exciting times ahead.
References
- Pennisi,E. (2007). Genetics. Working the (gene count) numbers: finally, a firm answer? Science. 316, 1113.
- Rana,T.M. (2007). Illuminating the silence: understanding the structure and function of small RNAs. Nat. Rev. Mol. Cell Biol. 8, 23-36.
- Gevaert,K., Van,D.P., Ghesquiere,B., and Vandekerckhove,J. (2006). Protein processing and other modifications analyzed by diagonal peptide chromatography. Biochim. Biophys. Acta. 1764, 1801-1810.
- Liu,H., Lin,D., and Yates,J.R., III (2002). Multidimensional separations for protein/peptide analysis in the post-genomic era. Biotechniques. 32, 898, 900, 902.
- Matallanas,D., Romano,D., Yee,K., Meissl,K., Kucerova,L., Piazzolla,D., Baccarini,M., Vass,K.O.E., Kolch,W., and O’Neill,E. (2007). RASSF1A elicits apoptosis through an MST2 pathway directing proapoptotic transcription by the p73 tumor suppressor protein. in press, Molecular Cell.
- Reyzer,M.L. and Caprioli,R.M. (2007). MALDI-MS-based imaging of small molecules and proteins in tissues. Curr. Opin. Chem. Biol. 11, 29-35.
Walter Kolch
The Beatson Institute for Cancer Research/ Institute for Biomedical and Life Sciences, University of Glasgow
Walter Kolch is the Director of Interdisciplinary Research Collaboration (IRC) Proteomics Technologies at the University of Glasgow. He began his work at Glasgow University in 2000, as the Professor of Molecular Cell Biology and in 2001 became the Director of the Sir Henry Wellcome Functional Genomics Facility. Along with contributing to a number of publications, his main achievements in terms of Proteomics research include; pioneering the use of dominant negative mutants to map signaling pathways in mammalian cells, developing biochemical and cellular drug screening assays crucial for the development of two widely used and highly specific PKC inhibitors, Goe6976 and Goe6815, co-discovering the crosstalk between the cAMP and Raf-1 signaling pathways and pioneering the use of proteomics for the mapping of signaling pathways. Mr Kolch is currently working on the development of enhanced proteomics technologies to understand cellular signaling networks and underpin rational approaches to discover and validate new drug targets, alongside characterisation of the metastasis suppressor function of RKIP and characterisation of new signaling pathways how Raf kinases regulate apoptosis and cell transformation. Further, Mr Kolch is also researching the role of protein Interactions in the regulation of signal transduction networks and the development of systems biology and computational modeling approaches to analyse cellular signal transduction networks.
Issue
Related topics
Related organisations
Beatson Institute for Cancer Research, University of Glasgow