Challenges in High-Content siRNA Screening
Posted: 28 November 2006 | | No comments yet
Huge progress has been made, both in RNA interference technology applied to mammalian cells and in automated microscopy to analyse gene functions upon silencing in the cellular context. Large-scale siRNA screens have been published recently, mainly applying assays that gain multi-parametric information on biological processes. It is a long way to establish an infrastructure that allows high-content siRNA screening, and in this article the major challenges are summarised.
Huge progress has been made, both in RNA interference technology applied to mammalian cells and in automated microscopy to analyse gene functions upon silencing in the cellular context. Large-scale siRNA screens have been published recently, mainly applying assays that gain multi-parametric information on biological processes. It is a long way to establish an infrastructure that allows high-content siRNA screening, and in this article the major challenges are summarised.
Huge progress has been made, both in RNA interference technology applied to mammalian cells and in automated microscopy to analyse gene functions upon silencing in the cellular context. Large-scale siRNA screens have been published recently, mainly applying assays that gain multi-parametric information on biological processes. It is a long way to establish an infrastructure that allows high-content siRNA screening, and in this article the major challenges are summarised.
RNA interference has revolutionised functional analysis of genes1,2. After the breakthrough demonstrated by the first application of chemically synthesised 21-meric short interfering RNAs (siRNA) in mammalian cells3, various methods of RNA interference have been established for application in mammalian cells that allow efficient and reproducible silencing of individual genes providing the opportunity to gain functional information on each individual gene.
Algorithms have been developed to predict siRNA molecules that exhibit a high probability of being functional. Such siRNAs are commercially available against all predicted genes of the human, mouse and rat genomes. Large libraries have been assembled to target individual classes of interest including the ‘druggable’ and the whole genome. Three technologies have been elaborated for gene silencing in mammalian cells:
- Chemically synthesised siRNAs3
- Short hairpin siRNAs (shRNA) expressed from vectors such as plasmids or viruses4,5
- Endoribonuclease-derived siRNAs (esiRNA) that are produced in vitro digesting double stranded long RNA by Dicer or RNaseIII6,7
Genome-wide libraries of all three types are available. A number of large-scale screening projects have already been accomplished and published8-16. Of particular interest to the pharmaceutical industry might be some recently published siRNA screens to sensitise existing chemotherapy currently in clinical practice17,18. This may help to identify new targets for drug discovery programmes or new combination treatments of existing drugs.
How to screen siRNA libraries and follow-up hits
A few lessons have been learned from large-scale experiments. Minimal requirements for studies applying RNA interference were set by Nature Cell Biology in 200319 and specified further for large-scale experiments in particular20-23. It is critical not only to elaborate an appropriate screening strategy, but also a hit follow-up strategy. This will verify the data generated by the primary screening and ensure the release of solid and reliable data. Beside variation in the assay, a major source of false-positives are ‘off-target effects’, meaning silencing of other targets than the desired, due to sequence homologies24-26. The desired assay must be properly validated applying appropriate negative and assay-specific positive controls and a reliable screening window, described in order to allow definition of ‘hits’. The rule to define a hit beyond the three times standard deviation from background achieved with the negative control might be applied, but dependent on assay specifications and hit rate that might be lowered also to twofold SD. More importantly, the hits must be correctly followed-up. A widely applied screening strategy is to apply 2-4 individual siRNA molecules per target in separate wells, rather than pools in triplicates, already for primary screening. To keep the costs low replicates might be skipped instead. In our experience at least three individual siRNAs should be applied, as two siRNAs per target do frequently end in a stand-off situation when only one molecule scores for the phenotype. This makes interpretation difficult. Results obtained at three out of three or two out of three siRNAs might be considered as real hits and followed up later while one out of three may be suspicious, therefore resulting from off-target activity. A second independent run will filter the initial hit list and only the overlap of the two runs might be taken further. Additionally, compared to expression profiling data of the cell line taken, a literature search will aid prioritisation. The selected hits should then be verified in secondary assays following an independent test principle. There are then a number of options available: (1) re-order the identical siRNA molecules, (2) order independent siRNAs against the same target, for example from a different vendor, or (3) apply another RNAi technology for hit verification such as short hairpin siRNAs or endoribonuclease-prepared siRNAs. For the solid hits, functionality of the molecules should then be demonstrated at mRNA level and/or at protein level if antibodies are available. The ultimate proof provides genetic rescue experiments. A very elegant method was described recently27,28: the mouse ortholog gene, including its upstream promoter sequence, was inserted into a bacterial artificial chromosome (BAC). This was then fused to a GFP and then stably integrated into the human cell line that had previously been applied for screening. Only the combined silencing of both genes should exhibit the observed phenotype while targeting the mouse or endogenous human gene individually by specific siRNAs. This should show no effect.
The next challenge towards implementing large-scale screens is to elaborate siRNA transfection protocols that are appropriate for high-thoughput. A broad range of mainly lipid-based siRNA delivery agents have been commercialised and serve well for easy-to-transfect cancer cell lines29. Unfortunately, the majority of interesting cell models is more difficult to transfect, thus requiring screening for an appropriate reagent and conditions. An interesting opportunity offers electroporation: A number of instruments have been marketed recently that allow transfection in a 96-well format. However, costs could be a severely limiting factor for this technology. For large-scale screening projects transfection must be automated, not only to ease the labour but to also help standardise the process and minimise variation.
The major challenges of High-Content Screening
Recent progress in instrumentation and image analysis development has made microscopy applicable to high-throughput screening. ‘High-Content Screening’ (HCS) is defined as multiplexed functional screening based on imaging multiple targets in the physiologic context of intact cells by extraction of multicolour fluorescence information30. For HCS the most modern automated microscopy systems provide up to 100.000 confocal images per day, at multiple colours, simultaneously and at an amazingly high resolution that allows for detailed analysis at sub-cellular levels. Image analysis tools provide multi-parametric pattern extraction and quantification.
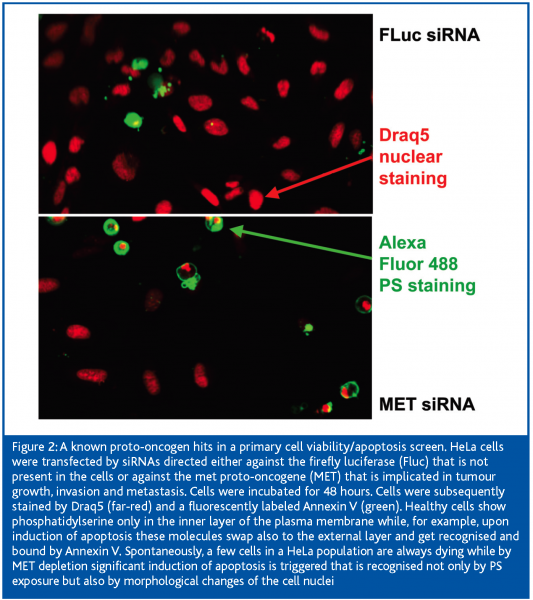
There are a number of advantages of high-content screening. Simultaneous staining in three or four colours allows extraction of various parameters from each cell in a qualitative and quantitative manner, such as intensity, size, distance or distribution. Cell-based assays reflect high physiological relevance and single cell analysis provides insight into the heterogenic response to any kind of treatment among a given cell population. The parameters might also be in correlation with each other. For example, any signal can be normalised against cell number upon nuclei staining and counting. On the other hand, in cellular assays higher variability is observed compared to biochemical assays. At least 200 cells should be covered, but that depends heavily on the type of assay and the frequency the desired phenotype occurs. Therefore, this number might be exceeded significantly.
A large number of image acquisition instruments are available. The most advanced ones provide confocal microscopy and are particularly designed towards high-throughput screening31 such as the OPERA (Evotec Technologies) or the InCell Analyser 3000 (GE Healthcare) or, at lower throughput, the Pathway BioImager (BD). Each of these three systems provide injection devices and incubation chambers with controlled temperature, CO2 concentration and humidity. The fourth confocal high throughput system, the ImageXpress Ultra (Molecular Devices), provides imaging by point scanning.
An essential component in the success of such high-content screening projects is the existence of sophisticated software algorithms that allow analysis of cellular fluorescence. The determination of morphological characteristics is to extract reliable and automatic information from the masses of captured images. Typically, nuclei are identified and masked first. The cell boundaries are then searched to mask the cell shape. Subsequently, the masks are laid over the image(s) of the other channel(s), and signals within the masks are measured. All mentioned automated microscopes are delivered with proprietary image analysis solutions for a broad range of biological events. For popular assays at cellular or sub-cellular level, such ready-to-use scripts are available and need only some fine-adjustments towards the cell line and/or conditions particularly used in one’s assay. Additionally, external commercial products are available or, alternatively, academic products such as the CellProfiler (http://www.cellprofiler.org/) might be used.
The key question is what to do with all the data. High-content screening can easily generate more than one Terabyte in both primary images and metadata. These must then be stored and managed per run. Process artefacts have to be identified and eliminated. Results are then compared between independent runs. After this, an appropriate hit verification strategy has to be applied, as discussed above. Finally, data will be compared to other internal and external data sources. Cluster analysis will assist in identifying networks and correlations.
What next?
Although tremendous progress has been achieved in HCS, there are major challenges still to be solved. More complex and physiologically relevant assays will be developed in primary cells, multi-cellular systems, organ cultures or whole model organisms. Large-scale kinetic screens to visualise complex dynamic processes in living cells have been implemented.32 Image analysis hardly follows the technical progress in image acquisition. Furthermore, bioinformatics will face new challenges bringing context to all of the data. Finally, linking functional (siRNA) and chemical screening will dramatically support systems biology and will broaden our understanding of complex biological processes.



References
- Tomari Y, Zamore PD: Perspective: machines for RNAi. Genes Dev. 2005 Mar 1;19(5):517-29.
- Dykxhoorn DM, Novina CD, Sharp PA: Killing the messenger: short RNAs that silence gene expression. Nat Rev Mol Cell Biol. 2003 Jun;4(6):457-67.
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T: Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001 May 24;411(6836):494-8.
- Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, Sheth N, Bradshaw J, Burchard J, Kulkarni A, Cavet G, Sachidanandam R, McCombie WR, Cleary MA, Elledge SJ, Hannon GJ: Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005 Nov;37(11):1281-8.
- Root DE, Hacohen N, Hahn WC, Lander ES, Sabatini DM: Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat Methods. 2006 Sep;3(9):715-9.
- Yang D, Buchholz F, Huang Z, Goga A, Chen CY, Brodsky FM, Bishop JM: Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. Proc Natl Acad Sci U S A. 2002 Jul 23;99(15):9942-7.
- Kittler R, Heninger AK, Franke K, Habermann B, Buchholz F.: Production of endoribonuclease-prepared short interfering RNAs for gene silencing in mammalian cells. Nat Methods. 2005 Oct;2(10):779-84.
- Aza-Blanc P, Cooper CL, Wagner K, Batalov S, Deveraux QL, Cooke MP: Identification of modulators of TRAIL-induced apoptosis via RNAi-based phenotypic screening. Mol Cell. 2003 Sep;12(3):627-37.
- Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heimerikx M, Kerkhoven RM, Madiredjo M, Nijkamp W, Weigelt B, Agami R, Ge W, Cavet G, Linsley PS, Beijersbergen RL, Bernards R: A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004 Mar 25;428(6981):431-7.
- Kittler R, Putz G, Pelletier L, Poser I, Heninger AK, Drechsel D, Fischer S, Konstantinova I, Habermann B, Grabner H, Yaspo ML, Himmelbauer H, Korn B, Neugebauer K, Pisabarro MT, Buchholz F: An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature. 2004 Dec 23;432(7020):1036-40.
- Pelkmans L, Fava E, Grabner H, Hannus M, Habermann B, Krausz E, Zerial M: Genome-wide analysis of human kinases in clathrin- and caveolae/raft-mediated endocytosis. Nature 2005, 436:78-86.
- Westbrook TF, Martin ES, Schlabach MR, Leng Y, Liang AC, Feng B, Zhao JJ, Roberts TM, Mandel G, Hannon GJ, Depinho RA, Chin L, Elledge SJ: A genetic screen for candidate tumor suppressors identifies REST. Cell. 2005 Jun 17;121(6):837-48.
- Kolfschoten IG, van Leeuwen B, Berns K, Mullenders J, Beijersbergen RL, Bernards R, Voorhoeve PM, Agami R: A genetic screen identifies PITX1 as a suppressor of RAS activity and tumorigenicity. Cell. 2005 Jun 17;121(6):849-58.
- Zhu C, Zhao J, Bibikova M, Leverson JD, Bossy-Wetzel E, Fan JB, Abraham RT, Jiang W: Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using RNA interference. Mol Biol Cell. 2005 Jul;16(7):3187-99.
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, Carpenter AE, Foo SY, Stewart SA, Stockwell BR, Hacohen N, Hahn WC, Lander ES, Sabatini DM, Root DE: A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 2006 Mar 24;124(6):1283-98.
- Espeseth AS, Huang Q, Gates A, Xu M, Yu Y, Simon AJ, Shi X-P, Zhang X, Hodor P, Stone DJ,Burchard J, Cavet G, Bartz S, Linsley P, Ray WJ, Hazudag D: A genome wide analysis of ubiquitin ligases in APP processing identifies a novel regulator of BACE1 mRNA levels. Mol Cell Neurosci. 2006 Sep 13; [Epub ahead of print].
- MacKeigan JP, Murphy LO, Blenis J: Sensitized RNAi screen of human kinases and phosphatases identifies new regulators of apoptosis and chemoresistance. Nat Cell Biol. 2005 Jun;7(6):591-600.
- Bartz SR, Zhang Z, Burchard J, Imakura M, Martin M, Palmieri A, Needham R, Guo J, Gordon M, Chung N, Warrener P, Jackson AL, Carleton M, Oatley M, Locco L, Santini F, Smith T, Kunapuli P, Ferrer M, Strulovici B, Friend SH, Linsley PS: siRNA Screens Reveal Enhanced Cisplatin Cytotoxicity in Tumor Cells Having Both BRCA Network and TP53 Disruptions. Mol Cell Biol. 2006 Sep 25 [Epub ahead of print].
- Editorial: Whither RNAi? Nat Cell Biol. 2003, 5:489-490.
- Krausz E, Grabner A, Kroenke A, Sachse C, Echeverri CJ: Optimising high throughput RNAi-based assays using transient transfection of synthetic siRNAs in cultured mammalian cells. In Engelke D.R. (ed): RNA Interference (RNAi)- Nuts & Bolts of RNAi Technology, p. 131-168, DNA Press LLC, (2003).
- Sachse C, Krausz E, Krönke A, Hannus M, Grabner A, Walsh A, Ovcharenko D, Dorris D, Trudel C, Sönnichsen B, Echeverri CJ: High throughput RNA interference strategies for target discovery and validation using synthetic short interfering RNAs: functional genomics investigations of biological pathways. Methods Enzymol. 2005, 392:242-277.
- Echeverri CJ, Perrimon N.: High-throughput RNAi screening in cultured cells: a user’s guide. Nat Rev Genet. 2006 May;7(5):373-84.
- Echeverri CJ, Beachy PA, Baum B, Boutros M, Buchholz F, Chanda SK, Downward J, Ellenberg J, Fraser AG, Hacohen N, Hahn WC, Jackson AL, Kiger A, Linsley PS, Lum L, Ma Y, Mathey-Prevot B, Root DE, Sabatini DM, Taipale J, Perrimon N, Bernards R.: Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat Methods. 2006 Oct;3(10):777-9.
- Semizarov D, Frost L, Sarthy A, Kroeger P, Halbert DN, Fesik SW.: Specificity of short interfering RNA determined through gene expression signatures. Proc Natl Acad Sci U S A. 2003, 100(11):6347-52.
- Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS.: Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003 Jun;21(6):635-7.
- Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, Fedorov Y, Baskerville S, Maksimova E, Robinson K, Karpilow J, Marshall WS, Khvorova A.: 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Methods. 2006 Mar;3(3):199-204.
- Kittler R, Pelletier L, Ma C, Poser I, Fischer S, Hyman AA, Buchholz F.: RNA interference rescue by bacterial artificial chromosome transgenesis in mammalian tissue culture cells. Proc Natl Acad Sci U S A. 2005 Feb 15;102(7): 2396-401.
- Poser I, Buchholz F: A new approach to specify RNAi experiments. Eur Pharm Rev 2006, 3:12-17.
- Ovcharenko D, Jarvis R, Hunicke-Smith S, Kelnar K, Brown D: High-throughput RNAi screening in vitro: From cell lines to primary cells. RNA. 2005, 11:985-993.
- Giuliano KA, DeBiasio RL Dunlay RT Gough A Volosky JM, Zock J, Pavlakis GN, DL Taylor DL:. High Content Screening: A new approach to easing key bottlenecks in the drug discovery process. J Biomol Screen. 1997, 2:249-259.
- Zemanová L, Schenk A, Valler MJ, Nienhaus GU, Heilker R: Confocal optics microscopy for biochemical and cellular high-throughput screening. Drug Discov Today 2003, 8:1085-1093.
- Neumann B, Held M, Liebel U, Erfle H, Rogers P, Pepperkok R, Ellenberg J.: High-throughput RNAi screening by time-lapse imaging of live human cells. Nat Methods. 2006 May;3(5):385-90.




