Cell-based imaging
Posted: 28 September 2006 | | No comments yet
Traditionally, in vivo histopathological findings have been used as part of standard readouts for safety assessment of new chemical entities (NCEs). The process typically involves dosing animals with NCEs for varying amounts of time, harvesting their major organs at the end of the study, preparing tissue sections and slides and visual examination of these slides by trained pathologists.
Traditionally, in vivo histopathological findings have been used as part of standard readouts for safety assessment of new chemical entities (NCEs). The process typically involves dosing animals with NCEs for varying amounts of time, harvesting their major organs at the end of the study, preparing tissue sections and slides and visual examination of these slides by trained pathologists.
Traditionally, in vivo histopathological findings have been used as part of standard readouts for safety assessment of new chemical entities (NCEs). The process typically involves dosing animals with NCEs for varying amounts of time, harvesting their major organs at the end of the study, preparing tissue sections and slides and visual examination of these slides by trained pathologists.
These studies can pinpoint target organ toxicity and other unexpected side-effects of drug candidates and as such, are prerequisites for drug regulators prior to initiating clinical testing in humans. However, these studies also consume relatively large amounts of time, labour, animals and chemical materials. As a result, they cannot routinely provide the necessary turnaround time for decision making in earlier phases of drug discovery and development. Recent advances in combinatorial library design, automated synthesis and high throughput screening have enabled increasing numbers of pharmacologically active compounds from diverse chemical spaces to be identified in a relatively short period of time. However, the quantity of each active compound is typically limited to less than 100 mg. Since the traditional animal studies require more compound and time, early toxicology screening approaches are greatly desired to prioritise the selection of chemicals for further development. In vitro screening approaches that are developed based on common mechanisms of drug-induced toxicity can fulfil this gap. Specifically, the in vitro correlate of histopathology, cellular imaging technology, has a special role to play to meet such demand.
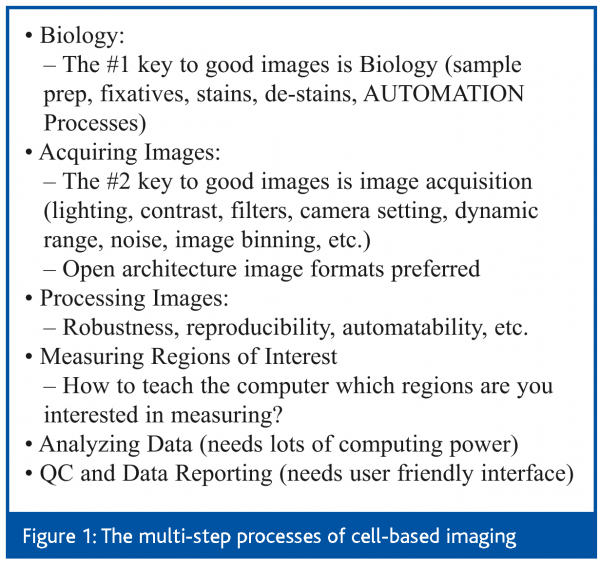
Cellular imaging technology provides an essential visual and quantitative tool to interrogate the pharmacological and toxicological effects of various types of perturbations to cells. This review will only focus on the toxicological effects of chemicals perturbations to cells, with a special emphasis on fluorescent microscopic imaging. Figure 1 depicts the multi-step processes of cell-based imaging.
One of the key enablers in recent history is the availability of image-quality, high-density microplates that are UV and far red transparent and allow efficient automatic signal focusing. Within these plates the multi-step processes of culturing and treating cells, fixing, washing, staining, de-staining and sample imaging can be performed. These steps are compatible with standard laboratory automation processes, bringing image-based assays to the mainstream of laboratory automation. The availability of fluorescent proteins that serve as endogenous sensors inside living cells (e.g., GFP/RFP/YFP families of fluorescent proteins), makes it possible to develop live cell and homogeneous assays with real-time imaging detection. When combined with a variety of fluorescent probes and stains, it is possible to quantitatively measure essentially any cellular compartments and macromolecules using fluorometric-morphometric, and spatial-temporal relationships1. In imaging hardware, the key enabler is the availability of entirely automated microscopy systems including those that keep the local environment compatible with normal cell physiology2. In imaging informatics, one can still use auxiliary hard-drives to archive images and perform visual characterization of these images on computer screens3. On the other hand, one can automate the entire image acquisition, archiving, analysis and data reporting steps to maximise the efficiency of screening and profiling chemical molecules3-5.
Apoptosis and Necrosis
One of the first major applications of cellular imaging in studying drug toxicity is in the differentiation of apoptosis and necrosis. Cellular morphology is the ‘gold standard’ in differentiating these two types of cell death. In general, necrotic cells swell and lyse, whereas apoptotic cells shrink and fragment. Chromatin in apoptotic cells forms electron-dense crescents at the nuclear envelope, then breaks up into ‘apoptotic bodies’ that may contain bits of chromatin6,7. In practice, however, differentiating apoptosis from necrosis can be difficult. Apoptotic cells may undergo secondary necrosis, during which they swell and lyse. In addition, the same chemical can elicit apoptosis at low doses and necrosis at higher doses and/or upon a longer duration of treatment. Because of these complexities, one cannot draw simple conclusions based on observations made at a single dose or time. The use of cellular imaging allows scientists to study apoptosis and necrosis morphologically on an individual cell-by-cell basis, and minimise false positive or false negative results from biochemical methods such as TUNEL8 and caspase activation9. In addition, observation of cellular morphology over time can provide additional insights in the time-sequence events leading up to apoptosis. For example, reduction in cell size was found to precede the appearance of phosphatidylserine on the cell surface10. In essence, the ability of image-based assays to simultaneously measure multiple early, intermediate and late apoptotic events provides far more information than single-parameter assays that provide only an ambiguous monovariate output11.
Micronucleus
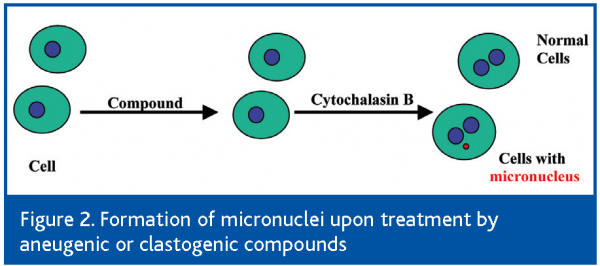
The micronucleus test is a genotoxicity test relevant for the risk assessment of cancer-inducing ability of a new chemical entity (NCE). A micronucleus (MN), or a small nucleus, is formed during the metaphase/anaphase transition of mitosis (or cell division). An MN may contain a whole piece of chromosome that arises from a whole lagging chromosome (aneugenic event leading to chromosome loss), or a partial piece of a whole chromosome that arises from a broken chromosome (clastogenic event) that does not integrate into the daughter nuclei. The combination of the micronucleus test with fluorescence in situ hybridisation (FISH) with a probe labelling the centromeric region of the chromosomes allows discrimination between micronuclei caused by aneugens (centromeric stain positive) and clastogens (centromeric stain negative)12. In addition, the cells that have completed mitosis and nuclear division vs. the cells that have not can be distinguished by culturing the cells with cytochalasin B, an inhibitor of actins13. The non-divided cells will thus have one large nucleus (mononucleated cells), and the divided cells will have two regular-sized nuclei (binucleated cells)12. The legitimate micronucleus is a small-sized nucleus present in the binucleated cells (Figure 2). Furthermore, the exclusions of apoptotic nuclei and necrotic nuclei are important for the micronucleus test. Therefore, the micronucleus test is a more challenging nuclear morphology test than the apoptosis vs. necrosis test. The MN test typically requires the examination and scoring of at least 2,000 cells per sample to reach statistical significance14. While this can be done visually under a microscope, it is tedious, time consuming and error prone, especially if a large number of compounds need to be assessed for many different treatment concentrations. Cellular imaging technology is well suited to increase the efficiency of such a test by automating the most rate-limiting steps. In one interesting study, manual scoring of the same sample by different operators resulted in 60% differences in micronucleus frequency, while automated scoring by image analysis algorithms is highly reproducible15. In another study using laser scanning cytometry (LSC), visual identification of MN combined with multiparameter measurement that took into account the DNA content and protein/DNA ratio made it possible to establish the gating parameters needed to exclude objects that were not true MNs. The capability of LSC to relocate MN for visual examination was also useful to confirm their identification16.
Phospholipidosis
Phospholipidosis, or phospholipid storage disorder, refers to an excessive accumulation of phospholipids in cells and tissues. Phospholipids are essential components of mammalian cell membranes, membrane protein complexes, and intracellular membranous complexes (phagosomes, lysosomes, mitochondria, etc.). Consequently, their synthesis, utilisation and turnover are highly regulated in mammalian cells. Phospholipidosis may occur as the result of metabolic and/or genetic disorders such as Niemann-Pick and Tay-Sachs disease, or may occur as a result of exposure to certain drugs17,18.
Cationic amphiphilic drugs can often induce phospholipidosis17. While a direct link between phospholipidosis and frank organ toxicity has not been established, phospholipidosis is often interpreted as a sign of drug accumulation and disturbance of phospholipid metabolism in affected tissues19. Therefore, for drug development, the implication is often the need for further studies to assess possible associated toxicities in longer-term in vivo studies, the reversibility of any toxicity after treatment cessation, etc. In a seminal study, primary hepatocytes were freshly isolated from human donors and plated into chamber slides. At the end of the drug treatment and phospholipid staining, the hepatocytes were examined under a laser scanning microscope and phospholipidosis was rated visually by a score of 0 to 4 (0 being no phospholipidosis and 4 being highly phospholipidotic20). Using this technique, the authors were able to attribute the observed phospholipidosis to a specific metabolite of the drug and were subsequently able to identify another drug candidate that did not induce phospholipidosis. In another study, a similar but more automated approach has been applied to monocytes and successfully differentiated drugs that induce phospholipidosis versus drugs that do not21.
Steatosis
Steatosis, or accumulation of neutral lipid droplets, can occur in all major tissues. In liver parenchymal cells (i.e., hepatocytes) it is referred to as ‘fatty liver’ and can occur in two forms: microvesicular and macrovacuolar. Microvesicular steatosis is characterized by tiny fat droplets that do not displace the nucleus. It can be induced by tetracycline, valproate, hypoglycin, several antiviral nucleosides, and a number of experimental toxins22. Macrovacuolar steatosis is characterised by large droplets of fat that displace the nucleus to the cell periphery. It can be induced by ethanol, methotrexate, and minocycline22. Again, the ‘gold standard’ way of differentiating the two types of steatosis is by examining the fat droplets under the microscope.
Using Nile red, a fluorescent dye used extensively to study fat accumulation in many types of cells (e.g., 23,24), steatosis can be imaged and quantified by epifluorescence microscopy. Several compounds known to cause hepatic fat accumulation in vivo were examined and most robustly increased Nile red staining in HepG2 cells. Required concentrations for increased Nile red staining were at least three-fold lower than the cytotoxic concentration determined by a resazurin reduction assay in the same cells24. In another study, qualitatively similar Nile red image results were obtained when primary canine or rat hepatocytes were used23.
Oxidative Stress and Cellular Thiols
Oxidative stress is an important mechanism of chemical-induced toxicity, and as such, a variety of intracellular redox probes have been developed to measure the level of oxidative stress in living cells(1,25-27). Acetaminophen (APAP) is the most widely used hepatotoxic drug on the market, causing 25% of all drug-induced liver injuries reported to hospitals. It has been known for a long time that APAP can undergo CYP2E1-mediated oxidation to N-acetyl-p-benzo-quinoemine (NAPQI), an electrophile that forms covalent adducts with glutathione and thiol groups on proteins. Using cellular imaging technology, it was revealed that in addition to the initial metabolic generation of NAPQI and thiol adducts, the resulting secondary oxidative stress may be more important in explaining APAP-induced liver injury28. In another example, low concentrations of cocaine treatment resulted in an increase in oxidative stress and enhancement in apoptotic cell death29.
Cellular thiols, especially glutathione, are important cytoprotective agents. Reduced levels of glutathione often parallel the induction of oxidative stress levels30. Intracellular levels of glutathione can be easily measured microscopically(31,32). Older literature used monochlorobimane and monobromobimane as GSH probes, but recent publications suggest that some of the newer fluorescent probes have better sensitivity and cellular retention33. While increases in oxidative stress and decreases in GSH levels may be measured by whole cell lysate assays, cellular imaging is more sensitive because it is able to measure subtle changes dynamically on a cell-by-cell basis, thus providing the chronology of toxin-induced injury to cells(34,35).
Mitochondrial dysfunction
Cellular imaging can also be used to image sub-cellular organelles such as mitochondria, lysosomes, the Golgi apparatus and cytoskeletal components. The commercial availability of organelle-specific probes make it possible to assess pathological changes at any sub-cellular organelle level36. Because of its central role in energy generation, the mitochondrion is an important target for chemical-induced toxicity(37-40). A variety of fluorescent probes responsive to mitochondrial membrane potential can be used for these imaging studies36.
Two key questions in a cellular imaging experiment are what probes to use and what image descriptors to collect. There are essentially two ways to think of this problem, one hypothesis-dependent and the other hypothesis-independent. In the former approach, one chooses probes and descriptors to look for specific biological effects such as micronucleus, steatosis, phospholipidosis, etc. In a hypothesis-independent approach, one would choose a broad range of probes and measure as many descriptors as possible for each probe. The latter approach is especially useful to detect unexpected or ‘off-target’ effects of drug actions3. In a landmark study of this type, Perlman et al. used automated microscopy to generate ~10^9 descriptor measurements in total. They then developed a statistical method to convert descriptor measurements into Z-scores for the differences between drug-treated cells and control cells. For data visualization, the authors used heat plots of Z-values versus drug concentration for each descriptor. Thus, drug effects (both targeted effects and ‘off-target’ side-effects) can be compared both visually and statistically41. Studies of this scale are only possible with the recent technological advances in fluorescence imaging, automated microscopy, and image informatics. Apart from experimental approaches, computational modelling of the imaging data can provide a theoretical framework to the inner workings of cells (e.g., 42). The combination of imaging data and computational modelling is also providing researchers a systems biology view of cellular responses to external perturbations43. The open microscopy initiative can further ensure that the image data will remain open and readable by future generations of image informatics tools regardless of image acquisition method and hardware, thus paving the ways for standardization and future compatibility44,45.
Conclusions
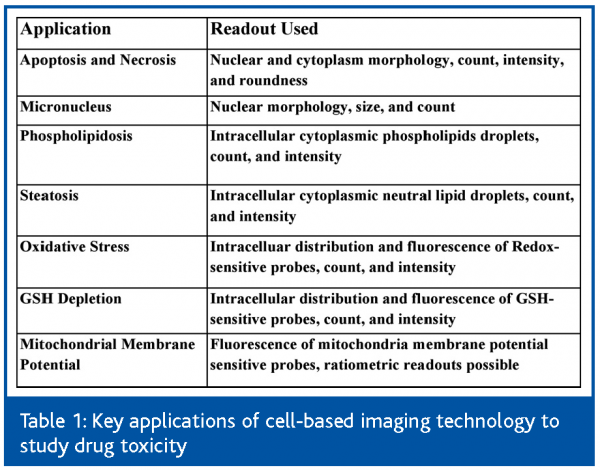
Recent advances in fluorescence biology, automated microscopy and image informatics make it possible to practice on a large scale, the morphologic screening of cells for both drug efficacy and toxicity. In addition, a panel of cell image-based assays has been developed, each targeting a known mechanism of drug toxicity (Table 1). The integrated usage of the automated cell-based imaging technology in drug discovery should enable the efficient design and selection of drug candidates with broader therapeutic indexes. The field of ‘Cytomics’46 has found its practical utility in drug discovery.
Acknowledgements
We wish to thank Dr. David de Graaf for support, helpful discussions and reviewing the manuscript. We also acknowledge valuable discussions from Drs. Peter O’Brien and Michael Banker. Space limitations prevented the citation of many important publications.



References
- Jameson, D.M., J.C. Croney, and P.D. Moens, Fluorescence: basic concepts, practical aspects, and some anecdotes. Methods Enzymol, 2003. 360: p. 1-43.
- Comley, J. and S. Fox, Growing market for high content analysis tools. Drug Discovery World, 2004. 5(2): p. 25-34.
- Mitchison, T.J., Small-molecule screening and profiling by using automated microscopy. Chembiochem, 2005. 6(1): p. 33-9.
- Abraham, V.C., D.L. Taylor, and J.R. Haskins, High content screening applied to large-scale cell biology. Trends Biotechnol, 2004. 22(1): p. 15-22.
- Giuliano, K.A., J.R. Haskins, and D.L. Taylor, Advances in high content screening for drug discovery. Assay Drug Dev Technol, 2003. 1(4): p. 565-77.
- Searle, J., J.F. Kerr, and C.J. Bishop, Necrosis and apoptosis: distinct modes of cell death with fundamentally different significance. Pathol Annu, 1982. 17 Pt 2: p. 229-59
- Wyllie, A.H., J.F. Kerr, and A.R. Currie, Cell death: the significance of apoptosis. Int Rev Cytol, 1980. 68: p. 251-306.
- Garrity, M.M., et al., Identifying and quantifying apoptosis: navigating technical pitfalls. Modern Pathology, 2003. 16(4): p. 389-94.
- Lovborg, H., J. Gullbo, and R. Larsson, Screening for apoptosis-classical and emerging techniques. Anticancer Drugs, 2005. 16(6): p. 593-9.
- Plasier, B., et al., Automatic image analysis for quantification of apoptosis in animal cell culture by annexin-V affinity assay. J Immunol Methods, 1999. 229(1-2): p. 81-95.
- Telford, W.G., A. Komoriya, and B.Z. Packard, Multiparametric analysis of apoptosis by flow and image cytometry. Methods Mol Biol, 2004. 263: p. 141-60.
- Kirsch-Volders, M., et al., The in vitro micronucleus test: a multi-endpoint assay to detect simultaneously mitotic delay, apoptosis, chromosome breakage, chromosome loss and non-disjunction. Mutat Res, 1997. 392(1-2): p. 19-30.
- Fenech, M., et al., HUMN project: detailed description of the scoring criteria for the cytokinesis-block micronucleus assay using isolated human lymphocyte cultures. Mutat Res, 2003. 534(1-2): p. 65-75.
- Kirsch-Volders, M., et al., Report from the in vitro micronucleus assay working group. Mutat Res, 2003. 540(2): p. 153-63.
- Frieauff, W., et al., Automatic analysis of the in vitro micronucleus test on V79 cells. Mutat Res, 1998. 413(1): p. 57-68.
- Smolewski, P., et al., Micronuclei assay by laser scanning cytometry. Cytometry, 2001. 45(1): p. 19-26.
- Halliwell, W.H., Cationic amphiphilic drug-induced phospholipidosis. Toxicologic Pathology, 1997. 25(1): p. 53-60.
- Reasor, M.J. and S. Kacew, Drug-induced phospholipidosis: are there functional consequences? Experimental Biology & Medicine, 2001. 226(9): p. 825-30.
- Hruban, Z., Pulmonary and generalized lysosomal storage induced by amphiphilic drugs. Environmental Health Perspectives, 1984. 55: p. 53-76.
- Gum, R.J., et al., Analysis of two matrix metalloproteinase inhibitors and their metabolites for induction of phospholipidosis in rat and human hepatocytes(1). Biochemical Pharmacology, 2001. 62(12): p. 1661-73.
- Casartelli, A., et al., A cell-based approach for the early assessment of the phospholipidogenic potential in pharmaceutical research and drug development. Cell Biology & Toxicology, 2003. 19(3): p. 161-76.
- Zimmerman, H., Hepatotoxicity: The adverse effects of drugs and other chemicals on the liver. 2nd ed. 1999, Philadelphia: Lippincott Williams & Wilkins.
- Amacher, D.E. and B.A. Martin, Tetracycline-induced steatosis in primary canine hepatocyte cultures. Fundamental & Applied Toxicology, 1997. 40(2): p. 256-63.
- McMillian, M.K., et al., Nile Red binding to HepG2 cells: an improved assay for in vitro studies of hepatosteatosis. In Vitro & Molecular Toxicology, 2001. 14(3): p. 177-90.
- Galati, G., et al., Idiosyncratic NSAID drug induced oxidative stress. Chemico-Biological Interactions, 2002. 142(1-2): p. 25-41.
- Amin, A. and A.A. Hamza, Oxidative stress mediates drug-induced hepatotoxicity in rats: a possible role of DNA fragmentation. Toxicology, 2005. 208(3): p. 367-75.
- James, L.P., P.R. Mayeux, and J.A. Hinson, Acetaminophen-induced hepatotoxicity. Drug Metabolism & Disposition, 2003. 31(12): p. 1499-506.
- Reid, A.B., et al., Mechanisms of acetaminophen-induced hepatotoxicity: role of oxidative stress and mitochondrial permeability transition in freshly isolated mouse hepatocytes. Journal of Pharmacology & Experimental Therapeutics, 2005. 312(2): p. 509-16.
- Diez-Fernandez, C., et al., Cocaine cytotoxicity in hepatocyte cultures from phenobarbital-induced rats: involvement of reactive oxygen species and expression of antioxidant defense systems. Biochemical Pharmacology, 1999. 58(5): p. 797-805.
- Kintzios, S., et al., Development of a novel, multi-analyte biosensor system for assaying cell division: Identification of cell proliferation/death precursor events. Biosens Bioelectron, 2005.
- Lantz, R.C., et al., Rapid reduction of intracellular glutathione in human bronchial epithelial cells exposed to occupational levels of toluene diisocyanate. Toxicological Sciences, 2001. 60(2): p. 348-55.
- Kurose, I., et al., Oxidative stress on mitochondria and cell membrane of cultured rat hepatocytes and perfused liver exposed to ethanol. Gastroenterology, 1997. 112(4): p. 1331-43.
- King, N., et al., A new method of quantifying glutathione levels in freshly isolated single superfused rat cardiomyocytes. Journal of Pharmacological & Toxicological Methods, 2004. 50(3): p. 215-22.
- Barhoumi, R. and R.C. Burghardt, Kinetic analysis of the chronology of patulin- and gossypol-induced cytotoxicity in vitro. Fundam Appl Toxicol, 1996. 30(2): p. 290-7.
- Thompson, D.C., R. Barhoumi, and R.C. Burghardt, Comparative toxicity of eugenol and its quinone methide metabolite in cultured liver cells using kinetic fluorescence bioassays. Toxicol Appl Pharmacol, 1998. 149(1): p. 55-63.
- Haugland, R.e.a., The Handbook — A Guide to Fluorescent Probes and Labeling Technologies. 10th ed. 2005, Molecular Probes.
- Lemasters, J.J., et al., Mitochondrial dysfunction in the pathogenesis of necrotic and apoptotic cell death. Journal of Bioenergetics & Biomembranes, 1999. 31(4): p. 305-19.
- Ding, W.X., H.M. Shen, and C.N. Ong, Critical role of reactive oxygen species and mitochondrial permeability transition in microcystin-induced rapid apoptosis in rat hepatocytes. Hepatology, 2000. 32(3): p. 547-55.
- Fellous, R., et al., Cytoplasmic accumulation of ditercalinium in rat hepatocytes and induction of mitochondrial damage. Cancer Research, 1988. 48(22): p. 6542-9.
- Santa Cruz, V., T.R. Dugas, and M.F. Kanz, Mitochondrial dysfunction occurs before transport or tight junction deficits in biliary epithelial cells exposed to bile from methylenedianiline-treated rats.[erratum appears in Toxicol Sci. 2005 Apr;84(2):418]. Toxicological Sciences, 2005. 84(1): p. 129-38.
- Perlman, Z.E., et al., Multidimensional drug profiling by automated microscopy. Science, 2004. 306(5699): p. 1194-8.
- Slepchenko, B.M., et al., Quantitative cell biology with the Virtual Cell. Trends Cell Biol, 2003. 13(11): p. 570-6.
- Valet, G., Cytomics, the human cytome project and systems biology: top-down resolution of the molecular biocomplexity of organisms by single cell analysis. Cell Prolif, 2005. 38(4): p. 171-4.
- Goldberg, I.G., et al., The Open Microscopy Environment (OME) Data Model and XML file: open tools for informatics and quantitative analysis in biological imaging. Genome Biol, 2005. 6(5): p. R47.
- Swedlow, J.R., et al., Informatics and quantitative analysis in biological imaging. Science, 2003. 300(5616): p. 100-2.
- Valet, G., Human cytome project, cytomics, and systems biology: the incentive for new horizons in cytometry. Cytometry A, 2005. 64A(1): p. 1-2.
Acknowledgement
This article first appeared in Issue 1, Volume 1 of American Drug Discovery




