A new approach to specify RNAi experiments
Posted: 23 May 2006 | | No comments yet
Large DNA-sequencing projects such as the Human Genome Project have provided the scientific community with a new challenge: to try to understand the information encoded in the primary sequence of the genome. Studies investigating the role and function of the components of the genome are often called functional genomics. These studies require a constant improvement […]
Large DNA-sequencing projects such as the Human Genome Project have provided the scientific community with a new challenge: to try to understand the information encoded in the primary sequence of the genome. Studies investigating the role and function of the components of the genome are often called functional genomics.
These studies require a constant improvement of tools that fulfil specific requirements. Recently, we have created a new powerful tool to improve functional genomic studies by combining two distinct methods: RNAi and BAC recombination technology.1
RNAi and its limits
RNA silencing or RNA interference (RNAi) is a formal description of the biological response to double-stranded RNA (dsRNA) initially discovered in the nematode Caenorhabditis elegans.2 By injecting dsRNAs into worms it was found that genes whose sequences were complementary to those of introduced dsRNAs were silenced.3 Subsequent work by many groups showed that the RNAi pathway, which is highly conserved in many organisms, is also present in mammals and most other eukaryotes. Briefly, in this pathway the RNase enzyme Dicer processes the dsRNAs into short interfering RNAs (siRNAs) of 21 to 28 nucleotides in lengths. These siRNAs are subsequently rearranged into a silencing complex called RISC (RNA-induced silencing complex) where they, now single-stranded, bind tightly to an Ago protein.4,5 Then the siRNA-containing RISCs identify complementary or nearly-complementary messenger RNA (mRNA) and mediate the cleavage of the target-mRNA (see review by Meister and Tuschl6).
In the same manner, that RNAi became a standard methodology for silencing the expression of specific genes in mammalian cells, the concerns about specificity and effects on unintended targets (off-target) increased. Several recent studies have shown that the specificity of silencing is not absolute. Transfection of dsRNA can activate the innate immune pathways including interferon-regulated responses that serve as antiviral mechanisms. Here, dsRNA activates the enzyme PRK (dsRNA-dependent protein kinase) resulting in sequence-independent destruction of RNA and a general repression of protein synthesis.7,8
Also off-target effects can occur if as little as 6 nucleotides of the siRNA are complementary to the 3´ UTR of a non-target mRNA.9,10 In addition, off-target effects at the level of protein synthesis without having corresponding effects on mRNA levels have been reported.11
Rules for effective RNAi experiments
It is therefore important to include control experiments to confirm the specificity of an RNAi phenotype. Four guidelines have been suggested to avoid misinterpreting results: 1. A good algorithm to design a siRNA with particular attention to the 5´ end of the antisense strand; 2. Several siRNAs against the same target mRNA should give the same phenotypic outcome, yet a negative control should not score. In addition, it is advisable to monitor the target protein levels in addition to the mRNA levels to accomplish discrimination between effective and ineffective siRNA; 3. Working with the lowest possible siRNA concentration; and 4. The performance of a rescue experiment where a version of the targeted gene, which cannot be recognised by the siRNA, reverts the phenotype.12
Even though rescue experiments are claimed to be the ultimate test of the specificity of an RNAi experiment, several hurdles have to be overcome. Silent point mutations can be introduced into a cDNA encoding the targeted gene, that destroy complementarities with the siRNA.13 This approach is limited by the availability of a full-length cDNA and an often complex cloning procedure. Furthermore, cDNAs do not allow the expression of alternatively spliced transcripts and due to their typically used artificial promoters will likely be overexpressed. Therefore, they mostly do not reflect physiological expression levels which may cause artificial effects.
We have developed a technology that circumvents the above-mentioned limits of cDNA and established bacterial artificial chromosomes (BACs) as rescue constructs for RNAi experiments in mammalian tissue culture cells.
BAC, YAC and PAC
During the last two decades, vectors containing complementary DNA (cDNA) constructs have been the key instrument for genetic engineering and studies of gene function. The introduction of cDNA/mRNA and subsequent protein expression in bacteria and eukaryotic cells can easily be achieved by cloning the desired construct into numerous vectors and plasmids available. Several cloning strategies allow modifications such as insertion of mutations, addition of markers etc. But these techniques are limited in some important ways: (a) the amount of DNA which can be inserted into high copy vectors is limited and inserts bigger than 10 kb are often difficult to obtain; (b) the expression of the cloned cDNA is typically controlled by a promoter derived from a viral or model vertebrate promoter that often does not reflect the physiological expression pattern of the gene. Therefore, the introduced gene is mis-expressed, interfering with the fine-tuned regulatory machinery in the cell. Artificial effects can occur which are difficult to interpret.
The results of the Human Genome Project showed that only 5% of the genome encode genes, but most of the regulatory elements are located in the intergenic DNA, the sequences between the coding sequence of the genes. These elements are responsible for the proper expression of a gene in time and space. In eukaryotic cells they are called expression domains and are believed to contain all elements necessary for correct gene expression.14-18
To carry out functional genomic studies it became necessary to include those cis-acting regulatory elements in special expression vectors. Artificial chromosome constructs such as bacterial artificial chromosomes (BACs), yeast artificial chromosomes (YACs) and P1 artificial chromosomes (PACs) serve this purpose well. They are characterised by much larger cloning capacities (ranging from 100 to 1000kb) than standard plasmids.18,19
By including the genomic sequence instead of using cDNA the expression of alternatively spliced transcripts is possible because all regulatory elements of a gene are present. Compared to YACs, especially BACs harbour some interesting features making them an ideal basis to study genes: (i) they possess high transformation efficiencies, (ii) form stable clones, and (iii) bacterial clones and libraries grow much faster than YACs.20 BACs are now also the preferred choice to generate transgenic animals.19,21
We have now demonstrated a new application of BACs – a reliable method to create RNAi-resistant transgenes by expressing murine bacterial artificial chromosomes in human cells. To facilitate selection of the cells expressing the transgene a selectable marker had to be placed onto the BAC, which was done by recombination.
BAC recombination
Conventional cloning methods greatly rely on the disposition of restriction endonuclease cleavage sites to ligate DNA molecules. But in long DNA fragments the frequent occurrence of cleavage sites limits the size of the DNA fragment that can be manipulated to less than ca. 20kb.22
DNA engineering strategies relying on homologous recombination to alleviate the limitations of the use of restriction enzymes and allow a wide range of DNA modifications at any chosen position and unlimited by fragment size (reviewed by Muyrers et al.23). A flexible assay for DNA manipulations on the basis of homologous recombination between linear and circular DNA has been developed in Escherichia coli. Here, the integration site is defined through homologous regions, which are stretches of DNA shared by the DNA molecules that recombine. For efficient recombination homologous arms of only 35 to 60 nucleotides are required.13 Since these regions can be selected at will, there is no limitation in the position of the modification. Thus, homologous arms are short enough to be made by oligonucleotide synthesis and are subsequently attached to the DNA fragment via polymerase chain reaction (PCR). This results in a linear PCR fragment, which exhibits homologous arms on both ends. Because homologous recombination events are rare, an antibiotic-resistant gene is included in the PCR fragment. Therefore, cells carrying the recombinant are easily identified through a selection procedure.
In a first round, a plasmid carrying the genes of the recombinases RecE and RecT is introduced into the cells. After inducing the L-Arabinose-sensitive promoter24, both proteins interact with each other and function via a double strand break repair mechanism25, whereas RecE is a 5’-3’ exonuclease and RecT a DNA annealing protein26. Since a pSC101 temperature-sensitive origin27 is used to express these proteins, a temperature shift simply eliminates their plasmids after side-specific recombination took place. The efficiencies achieved with this so called ‘ET cloning’ or ‘recombineering’ are consistently high and typically more than 80% of the candidates are correctly recombined.25
BACs and RNAi rescue experiments
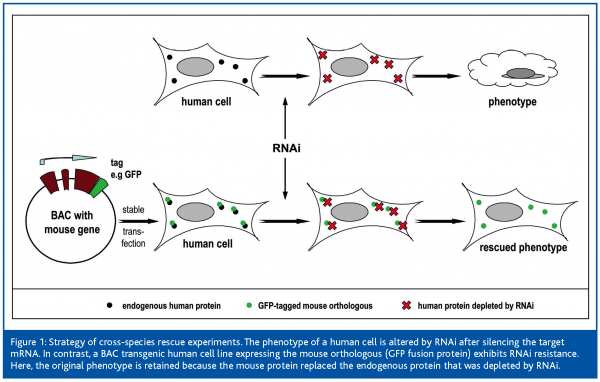
A successful rescue experiment is based on the fact that an RNAi resistant version of the target gene is expressed in the cells, reversing the induced phenotype. We tested a new approach by introducing BACs encoding orthologous genes from a closely related species. RNAi resistance of these transgenes was achieved by using BACs encoding mouse not human genes. Mouse homologous are currently known for more than 99% of the human genes.28 They exhibit large homologies to human proteins, but sufficiently differ on DNA levels. Therefore, in a cell that additionally expresses a mouse orthologous, RNAi will only run down the human transcript. Mouse proteins are very likely to function in a human environment and hence they are able to replace a human protein depleted by RNAi. Furthermore, this cross-species strategy confers RNAi resistance to the transgenes without the need to introduce point mutations.
In order to allow stable integration of the BAC into mammalian cells we developed a selection cassette (kanamycin/neomycin) to be used in Escherichia coli as well as in mammalian cells. Subsequently, these modified BACs were transfected into HeLa cells and stable cell clones were selected. Expression of the murine transgene at physiological levels and existence of alternative spliceforms were confirmed by RT-PCR. Then, RNAi experiments using esiRNA (endoribonuclease-prepared siRNA29) were carried out on essential genes. RT-PCR showed that there was a strong reduction of the human-specific product compared with the mouse-specific product. Furthermore, BAC-transgenic HeLa, transfected with esiRNA targeting an essential human transcript, grew essentially like wildtype HeLa as well as HeLa transfected with a negative control (firefly luciferase). In contrast, transfection of the same esiRNA into wild-type HeLa resulted in a markedly lower cell density and many dead cells. Further analysis of the mitotic spindle morphology revealed, that the spindle defect, caused by the esiRNA used, was reverted in the BAC-transgenic HeLa (see Figure 1).
This concluded that the introduced transgene was able to rescue the RNAi-induced phenotype. In principle, this concept should be applicable not only to human and mouse, but also to other closely related species, e.g. Drosophila and nematode strains.
However, it is important to consider that even through less than 1% of the human genes will not have any orthologous in the mouse genome, there is a risk that a cross-species BAC will not rescue. In these cases, the RNAi target region or the 3´ UTR of a BAC from the same species needs to be modified by point-mutagenesis.30
A new era
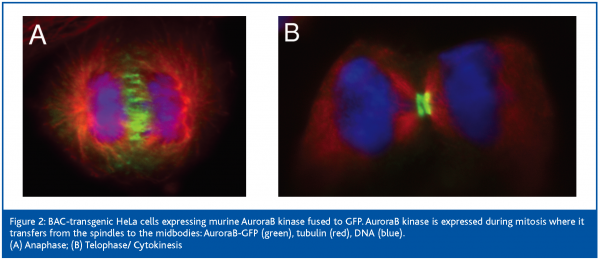
We have provided a straightforward way to test specificity of RNAi experiments by establishing BACs as rescue constructs in mammalian tissue culture cells. Using this technology will help to probe the specificity of RNAi experiments and hence will make these experiments more reliable. A simple variation of our approach led to further applications: instead of using BAC recombination to insert a selectable gene in the BAC backbone, fusion proteins carrying fluorescence markers (e.g. GFP) followed by a selectin marker cassette, were generated. These fusion proteins can now be used in localisation studies, function analysis and to purify protein complexes (Figure 2).
Again, the physiological expression of the transgenes will avoid artefacts of mis-localisation of the fusion proteins, or co-purification of proteins complexes, that do not normally form under physiological conditions. Using this approach in combination with RNAi also allows the sole expression of the tagged transgene, hence mimicking homologous recombination. With the full development of these technologies and its scaling to high throughput generation of BAC transgenic cell lines, a new era in mammalian functional genomics may just be around the corner.


References
- Kittler R, Pelletier L, Ma C, Poser I, Fischer S, Hyman AA, Buchholz F. 2004. RNA interference rescue by bacterial artificial chromosome transgenesis in mammalian tissue culture cells. Proc. Natl. Acad. Sci.102, 2396-2401
- Fire A, Albertson D, Harrison SW, Moerman DG.1991. Production of antisense RNA leads to effective and specific inhibition of gene expression in C. elegans muscle. Development. 113, 503-514
- Fire A et al. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 391, 806-811
- Hammond SM, Bernstein E, Beach D, Hannon GJ. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 404, 293-296
- Martinez J, Tuschl T. 2004. RISC is a 5´ phosphomonoester-producing RNA endonuclease. Genes Dev. 18, 975-980
- Meister G, Tuschl T. 2004. Mechanisms of gene silencing by double-stranded RNA. Nature. 431, 343-349
- Williams BR. 1997. Role of double-stranded RNA-activated protein kinase (PRK) in cell regulation. Biochem Soc Trans. 25, 509-513
- Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR. 2003. Activation of the interferon system by short-interfering RNAs. Nature Cell Biol. 5, 834-839
- Jackson AL, Linsley PS. 2004. Noise amidst the silence: off-target effects of siRNAs?. Trends Genet. 20, 521-524
- Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, Fedorov Y, Baskerville S, Maksimova E, Robinson K, Karpilow J, Marshall WS, Khvorova A. 2006. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nature Methods. 3, 199-204
- Scacheri PC, Rozenblatt-Rosen O, Caplen NJ, Wolfsberg TG, Umayam L, Lee JC, Hughes CM, Shanmugam KS, Bhattacharjee A, Meyerson M, Collins FS. 2004. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc Natl Acad Sci USA. 101, 1892-1897
- Hannon GJ, Rossi JJ. 2004. Unlocking the potential of the human genome with RNA interference. Nature. 431, 371-378
- Zhang Y, Buchholz F, Muyrers JPP, Stewart AF. 1998. A new logic for DNA engineering using recombination in Escherichia coli. Nature Genet. 20, 123-128
- Elgin SC. 1990. Chromatin structure and gene activity. Curr Opin Cell Biol. 2, 437-445
- Laemmli UK, Kas E, Poljak L, Adachi Y. 1992. Scaffold-associated regions: cis-acting determinants of chromatin structural loops and functional domains. Curr Opin Genet Div. 2, 275-285
- Dillon N, Grosveld F. 1994. Chromatin domains as potential units of eukaryotic gene function. Curr Opin Genet Dev. 4, 260-264
- Bonifer C. 2000. Developmental regulation of eukaryotic gene loci: which cis-regulatory information is required?. Trends Genet. 16, 310-315
- Montoliu L. 2002. Gene transfer strategies in animal transgenesis. Cloning Stem Cells. 4, 39-46
- Giraldo P, Montoliu L. 2001. Artificial chromosome transgenesis in pigmentary research. Transgenic Res. 10, 83-103
- Frijters ACJ, Zhang Z, van Damme M, Wang G-L, Ronald PC, Michelmore RW. 1997. Construction of a bacterial artificial chromosome library containing large EcoRI and HindIII genomic framents of lettuce. Theor Appl Genet. 94, 390-399
- Magdaleno SM, Curran T. 1999. Gene dosage in mice – BAC to the future. Nat Genet. 22, 319-320
- Ioannou PA, Amemiya CT, Garnes J, Kroisel PM, Shizuya H, Chen C, Batzer MA, de Jong PJ. 1994. A new bacteriophage P1-derived vector for the propagation of large human DNA fragments. Nature Genet. 6, 84-89
- Muyrers JPP, Zhang Y, Stewart AF. 2001. Techniques: Recombinogenic engineering – new options for cloning and manipulating DNA. Trends Biochem Sci. 26, 325-331
- Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 177, 4121-4130
- Muyrers JP, Zhang Y, Stewart AF. 2000. ET cloning: Think recombination first. In Genetic Engineering (NY), Principles and Methods (Vol. 22) (Setlov, JK, ed.), 77-98
- Kolodner R, Hall SD, Luisi-DeLuca C. 1994. Homologous pairing proteins encoded by the Escherichia coli recE and recT genes. Mol Microbiol. 11, 23-30
- Hashimoto-Gotoh T, Sekiguchi M. 1977. Mutations of temperature sensitivity in R plasmid pSC101. J Bacteriol. 131, 405-412
- Waterston RH et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature. 420, 520-562
- Yang D, Buchholz F, Huang Z, Goga A, Chen CY, Brodsky FM, Bishop JM. 2002. Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. Proc Natl Acad Sci USA. 99, 9942-9947
- Yang Y, Sharan SK. 2003. A simple two-step, ´hit and fix´ method to generate subtle mutations in BACs using short denatured PCR fragments. Nucleic Acids Res. 31, e80




