The human plasma proteome: A biomarker pool too deep to explore?
Posted: 23 May 2006 | | No comments yet
The human blood plasma harbors treasure, which, like most treasures, is not easily attained, and finding it requires ingenuity, endurance and possibly a grain of luck. The blood plasma is the largest (most proteins) and deepest (widest dynamic range) of the human proteomes. In order to ‘triumph over’ it, it is necessary to overcome an enormous protein concentration range to finally be rewarded with the possible discovery of biomarkers. But is this a realistic challenge we are facing or is this plasma pool too deep to explore?
The human blood plasma harbors treasure, which, like most treasures, is not easily attained, and finding it requires ingenuity, endurance and possibly a grain of luck. The blood plasma is the largest (most proteins) and deepest (widest dynamic range) of the human proteomes. In order to ‘triumph over’ it, it is necessary to overcome an enormous protein concentration range to finally be rewarded with the possible discovery of biomarkers. But is this a realistic challenge we are facing or is this plasma pool too deep to explore?
The human blood plasma harbors treasure, which, like most treasures, is not easily attained, and finding it requires ingenuity, endurance and possibly a grain of luck. The blood plasma is the largest (most proteins) and deepest (widest dynamic range) of the human proteomes. In order to ‘triumph over’ it, it is necessary to overcome an enormous protein concentration range to finally be rewarded with the possible discovery of biomarkers. But is this a realistic challenge we are facing or is this plasma pool too deep to explore?
The human blood plasma proteome
A closer look at human plasma reveals that there are both advantages and disadvantages in working with it. Like urine and saliva, it is easily collectable, and the fact that it likely contains proteins from nearly all the cells in the body makes it an especially interesting source for discoveries with diagnostic application. However, due to the wide range of concentrations in this vast plasma protein pool, it can be extremely difficult to detect low abundance proteins, which most biomarkers are expected to be – it is analogous to finding a needle in a haystack. 99% of plasma protein content is accounted for by only 22 proteins which include albumin, transferrin, fibrinogen, and others. Among the remaining 1% are the proteins of interest, given that the hypothesis is true that many promising plasma biomarkers are indeed secreted proteins. To make things even more complicated, besides harboring true plasma proteins, which are proteins that exert their function within the plasma, the plasma also contains proteins and protein fragments from dead or dying cells, and cells undergoing remodeling. Detailed knowledge of the content of the plasma is required if we are to distinguish whether a cellular protein found in the plasma is the result of apoptosis for example, or due to a disease state or process. The detection of a tumor biomarker can be complicated by the temporally dynamic nature and the large dilution volume of plasma, depending on the sampling location. The human body contains approximately 12 liters of extracellular fluid, 2.5 liters of which is plasma. Any sample taken represents a snapshot in time. Moreover, proteins with a molecular mass of < 45 kDa are rapidly cleared by the kidneys. For instance, the half-life of albumin (MW 69 kDa) is 15-19 days, while that of haptoglobin (45 kDa) is only 2 days(1). Drug molecules usually have an average plasma half-life of a few hours. Of course, plasma half-lives are not merely dependent on MW but also on the degree of albumin-binding and on physical factors including glycosylation rate, pH, and hydrophilicity. Increased levels of a plasma biomarker in patients with impaired renal function could be due to decreased renal clearance, increased pathological state, or both, but often the concern is that rapid excretion may reduce the concentration of low mass biomarkers below detection limits.
Characterization of the plasma proteome is therefore not easy. In addition to continuous instrumentation and technology refinement, investigators must be open to and able to manage new methodologies and techniques, as well as possess the knowledge and ability to handle and validate the vast amount of data generated. The goal is to define and establish a reliable normal human blood plasma reference dataset to create a solid foundation for future biomarker discoveries.
A blood plasma protein reference set
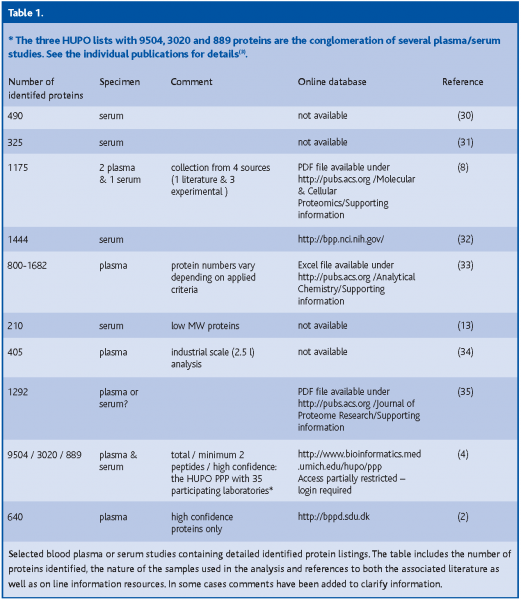
When considering mass spectrometry (MS)-based proteomics studies, it appears as if some large scale proteomics projects are more concerned with the generation of large protein lists rather than in concentrating on the quality of the data contained within. In a recent report, we have taken the stance that quality is more important than quantity and we have chosen to report only those proteins of very high confidence(2). A list such as this is indispensable for proteomics experiments where the goal of the study is biomarker discovery.
The Plasma Proteome Project (PPP) undertaken by the Human Proteome Organization (HUPO) underscores the need for a baseline dataset reflecting the true content of normal healthy human plasma. The PPP consisted of 35 laboratories from 13 countries, and included a bioinformatics group to organize the data. The PPP addressed questions regarding the best technology platform for the characterization of plasma and serum proteins, investigating such factors as the influence of specimen collection and handling procedures, whether the most abundant plasma proteins should be depleted, and whether anti-protease cocktails are desirable(3).
Besides evaluating technical and methodological variables, the international nature of the PPP also made it possible to address questions regarding potential differences in plasma or serum based on ethnicity(4,5,6) and gender(7). More comparative work is needed to reveal possible variations based on ethnicity, gender, age, and the effect of smoking. Confirmed divergence would emphasize the need for multiple plasma reference data sets.
The PPP yielded 9504 plasma/serum proteins, of which 889 proteins were rated as high confidence(4).
To complement the efforts of the PPP we also employed various techniques for depletion and pre-fractionation to deal with the enormous complexity and protein concentration range of plasma, coupled with the use of high-resolution instrumentation and consistent data treatment. We established a reference set of plasma proteins in which we are very highly confident(2).
The difficulty in establishing the true protein content of the human plasma is exemplified by very little overlap between independent datasets. Anderson et al.(8) investigated the congruence between plasma proteins derived from four different sources. One source was the literature, while the three others were MS-based and had the immunoglobulins (Ig) removed. A combined total of 1,175 proteins were identified from the 4 sources and of these, 980 occur in only 1 source. 195 proteins were found in at least 2 of the 4 datasets, 102 proteins were identified in exactly 2, and 47 proteins were identified in 3 datasets. Only 46 proteins were found in all 4 datasets. Our reference set identifies all 46 proteins found in those 4 sources, and 41 of the 47 proteins found in 3 datasets as well. An International Protein Index (IPI) accession numbers-based comparison between the HUPO high confidence list and our high confidence list revealed 278 proteins that were found to be in common.
All of these experiments employed a broad spectrum of sample handling and analysis techniques and importantly, the validation criteria applied varied considerably among different studies. Clearly, repetition of scientific experiments using different samples is a basic and vital scientific premise to corroborate data, since the repeated identification of a false positive is less likely as the number of repetitions increases. Even though the massive amount of data obtained from a single experiment already prohibits the feasibility of manual data validation, we still find it necessary to ensure accuracy. Unfortunately, search engines like MASCOT or SEQUEST are not currently sophisticated enough to negate the need for manual confirmation of proteomic data identification.
The investigations undertaken by the PPP and us clearly demonstrate the difficulty in identifying a reliable plasma protein dataset that can be used as a reference, where the vital prerequisites of comparable technology, analysis tools, sample handling and data validation standards have been filled. It is reasonable to expect a certain degree of variation in the protein profile identified between MS runs. Estimates of the variation attributable to technical replication issues average 24%(9,10). Biological variation has been estimated to be from 24-70%(10).
In summary, the work of the PPP and our laboratory were important steps in attempting to establish a high-confidence blood plasma protein reference set but more studies are needed. The results suggest the use of plasma rather than serum(4,11) and the employment of anticoagulation substances such as EDTA or citrate(4,6). Combinations of techniques such as the depletion of high abundance proteins together with plasma pre-fractionation prior to MS/MS analysis are helpful in overcoming plasma protein complexity and concentration range issues, but the loss of small proteins that are bound to high abundance proteins still presents a problem. A listing of different plasma proteomes is available (Table 01).
Preparative and analytical methodologies and instrumentation
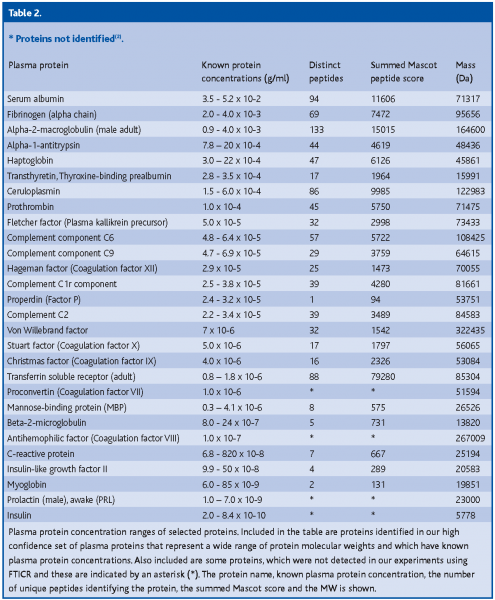
The MS methodologies and technologies currently employed cannot yet cover the 9-12 log portion of the plasma protein concentration range (~10-2 –10-12 g/ml). In our study we observed that despite the extremely high dynamic range covered by the Fourier Transform Ion Cyclotron Resonance (FTICR) detector, the overall plasma protein concentration range currently resolvable is only up to 7 orders of magnitude (10-2 – 10-9 g/ml) (Table 02). We were able to comprehensively cover up to 4 orders of magnitude (10-2 – 10-5/10-6 g/ml). The less abundant a protein is and the smaller its MW, the more unlikely is its detection, but this does not exclude the fact that some larger proteins were also not identified in each run. For instance, the precursor of anti-hemophilic factor (coagulation factor VIII) was not detected despite its large molecular weight (MW) of 267 kDa, while proteins that are known to be 1 to 2 orders of magnitude less abundant and with much smaller MWs, such as C-reactive protein and myotrophin, were conclusively identified.
Hence, prior to conducting comparative analysis to identify disease-specific biomarkers, it is a prerequisite that MS-based proteomic technology must be capable of providing the necessary protein concentration range. Researchers are investigating alternative strategies that could reduce the complexity of plasma, and mass spectrometer manufacturers are continuously striving to improve the instrumentation.
The Plasma complexity issue is typically addressed by employing techniques for serum or plasma depletion of high-abundance proteins, followed by multiple fractionation steps prior to MS-based analysis. A recent review on sample depletion and pre-fractionation is available(12).
The depletion of high abundance plasma proteins is commonly carried out using two key methods: resin-based (e.g. Cibacron Blue) and antibody-based (multiple affinity removal system (MARS), IgY-microbeads) removal. However, major well-founded concerns exist regarding these depletion practices because they concomitantly remove potentially important low molecular weight, low abundance proteins such as cytokines or hormones which may have diagnostic potential. Zhou et al.(13) used a series of antibodies against high abundance proteins to capture these as well as the smaller proteins associated with them. More than 200 associated proteins were found, including some currently utilized biomarkers such as PSA (prostate cancer), and some candidate molecules. This study confirmed that small MW proteins, including some biomarkers, are bound to large circulatory proteins and this helps prevent the smaller molecules from being rapidly cleared by the renal system. The selective removal of proteins such as albumin but not the albumin-bound proteins, as well as the recovery of low abundance proteins during the depletion steps, is desired(12).
In our hands the depletion of albumin from plasma allowed for the identification of a substantially higher number of validated proteins as well as a significant increase in the proportion of identified small MW proteins(2). Albumin depletion thus seemed to ‘unmask’ smaller proteins by lowering the effective protein concentration range.
Interestingly, the depletion of six high abundance proteins, including albumin, transferrin, haptoglobin, alpha-1-antitrypsin, IgA and IgG did not significantly increase the number of proteins identified compared to a similar experiment without depletion; on the contrary, their depletion resulted in the loss of proteins in the small MW range(2).
Following depletion, investigators agree that fractionation of plasma or serum prior to further analysis is a must and many fractionation strategies exist. In addition to the standard 1D- and 2D-PAGE, classical chromatography (i.e. reverse phase, RP; strong cation exchange, SCX), and OGE (Off-gel-isoelectric focusing/pI), membrane based, preparative electrophoresis based on size and charge, microscale solution phase IEF, free flow electrophoresis (FFE) based on size and pI, and liquid-based chromatofocusing followed by non-porous reverse phase chromatography based on pI and hydrophobicity are used.
In our study, a comparison between plasma separation by 1D-PAGE and OGE showed that the 1D PAGE-separation was at least as effective, if not more so, than the OGE technique regarding the number of proteins identified(2). The use of OGE seemed to adversely affect the identification of proteins in the 10-15 kDa range, implying that plasma separation by OGE may be less ideal for the identification of small MW or low abundance proteins.
At present, several independent serial depletion and separation strategies are required to obtain a more comprehensive picture of the human plasma proteome, and there is not yet any single or combination of analytical platforms that can deliver comprehensive, proteome- wide coverage.
Sample preparation limitations pose significant technological challenges for new technological developments, including new mass spectrometry instrumentation. Matrix-assisted-laser-desorption time-of-flight (MALDI-TOF) MS is currently very popular and one of the most accessible techniques. The introduction of chromatographic magnetic beads to MALDI-TOF analysis makes the usual off-line sample processing compatible with automation, enabling high sample throughput and maximum reproducibility. The use of a commercial kit employing magnetic beads for proteomic and biomarker studies is currently under scrutiny(14).
Surface-enhanced laser desorption ionization (SELDI)-TOF, an extension of the MALDI process of sorts, has been increasingly applied as a platform for serum/plasma biomarker discovery. A subset of plasma proteins preferentially binds to a chip, based on the surface chemistry (reverse or neutral phase, anionic or cationic exchange, or metal affinity) of the chip, allowing for sample processing in situ and reducing procedural losses. Furthermore, the utilization of different chip surface chemistry selectively eliminates high abundance proteins that may mask the subtle signals of lower abundance proteins. Simultaneous sub-fractionation is possible with Ciphergen’s Protein Chip Array®. In addition to criticism regarding its reproducibility, SELDI suffers from limited protein binding capacity and surface-derived mass spectrometric noise.
A revolutionary breakthrough in the study of biological macromolecules was recognized in 2002 with the shared Nobel Prize in Chemistry for soft laser desorption (SLD, in the form of MALDI)(15), and electrospray ionization (ESI) mass spectrometry(16,17). Capillary liquid chromatography has been interfaced with electrospray ionization (ESI)-ion trap MS and recently also with ESI-Fourier FTICR MS. The linear ion trap (LTQ) Orbitrap which appeared in 2005 represents the first completely new mass analyzer technology to be introduced to the market in over 20 years.
The LTQ is capable of detecting MS and MSn spectra at very high sensitivity, while the FTICR possesses a superior dynamic range and mass accuracy. Incidentally, our plasma analysis employed a hybrid LTQ-FTICR mass spectrometer system and to further enhance the reliability of our data, we employed MS3(2).
The LTQ-orbitrap has recently been tested(18). Preliminary results show similar mass accuracies with absolute mass errors of 0.6/0.7 ppm and 0.5 ppm for the FT-ICR and the LTQ-orbitrap, respectively. High mass accuracy in these instruments is achieved differently (lock mass for the LTQ-orbitrap and selected ion monitoring (SIM) scans for the FTICR), and employment of the LTQ-orbitrap saves time because the isolation and injection of peptides of a narrow mass range into the ICR (SIM scans) can be omitted.
The exceedingly robust and highly accurate masses obtained with both instruments constrain peptide candidates to just a few sequences, thereby greatly facilitating peptide identification and thus virtually reducing the problem of false positive peptide identification to zero(18). High mass accuracy also assists with the identification of post-translational modifications, which can be very informative. For example, the loss of the BRACA1 gene is often the cause for breast cancer, while post-translational phosphorylation at critical sites is essential for its tumor-suppressing function. Furthermore, the accurate mass determination of plasma proteins may allow for identification of candidate biomarkers identified by techniques such as protein profiling.
An alternative viewpoint is that higher sensitivity and mass accuracy are not necessarily the best choices for biomarker discovery. Instead, a more simplistic time of flight instrument is preferred which tends to be relatively more quantitative and thus is more reproducible for protein expression profiling(19).
Despite some controversy, most scientists agree that proteomics will ultimately make it to the clinic. However, mass spectrometry-based proteomics is seen as a discovery technique rather than as a diagnostic tool by some, and mass spectrometers are considered to be too costly for clinical laboratories. Although this may be largely true for the present and there are not yet any ‘clinical grade’ MS instruments, this situation is very likely to change as manufacturers are motivated to develop a cost-effective platform for clinical diagnostics.
Biomarker discovery
A biomarker is simply a protein whose presence in a biological fluid, such as blood plasma or urine, signals disease. The inherent property of a biomarker lies in its ability to provide an early indication of disease progression, the detection of recurrence, and/or the identification of high-risk individuals. The clinical imperative for the discovery of biomarkers and the development of diagnostic and prognostic tests derives from the significant impact that diseases such as cancer have when undiagnosed and untreated, and/or when found at an advanced stage.
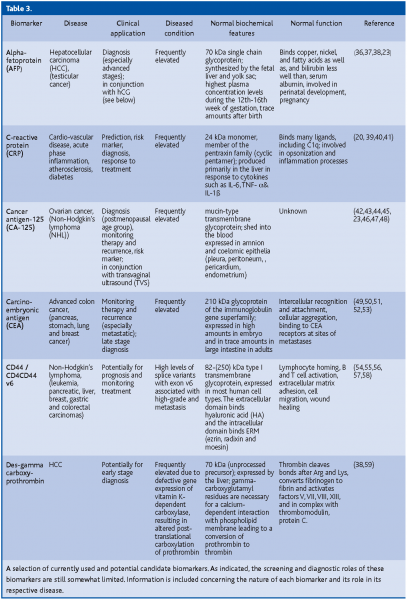
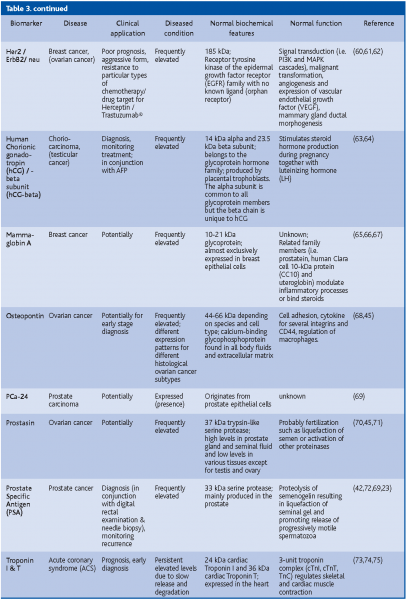
A putative biomarker is C-reactive protein (CRP). A growing number of studies have shown that high levels of CRP consistently predict new coronary events in patients with unstable angina and acute myocardial infarction (heart attack). Higher CRP levels are also associated with a lower survival rate of these people, and may indicate an increased risk that an artery will recluse after it has been opened by balloon angioplasty. High levels of CRP in the blood appear to be correlated with the prognosis and recurrent events in patients with stroke and peripheral arterial disease(20,21,22). So far, the biomarker with the highest sensitivity (>90%) and which has been successfully used for early disease detection is PSA. However, because of its low specificity (25%)(23) it is best used in conjunction with medical examination (digital rectal exam for prostate cancer) (Table 3).
The traditional way of discovering biomarkers is through the identification of a single biomarker species in a sample. Assuming that promising plasma/serum biomarkers are secreted proteins or extracellular domains that have been shed into the plasma, LC-MS/MS is most commonly employed to identify proteins that are unique or of higher abundance in samples obtained from patients with a disease, compared to samples from a healthy control.
However, some scientists see the future in biomarker patterns, using protein chips and pattern recognition software to probe serum or plasma for biomarkers in the form of a series of mass spectrometric peaks representing unidentified peptides.Petricoin and Liotta(24) published proteomic profiles that could be used to diagnose early ovarian cancer. The characterization of each peak in a biomarker panel is considered by some as a hindrance to early disease detection. Other investigators are concerned and think that without the knowledge of the identity of the proteins constituting the pattern, the method will remain empirical and difficult to validate, reproduce, standardize and quality control(25,26,27). An attempt to reproduce the results using Petricoin and Liotta’s own raw dataset has so far been unsuccessful(28). Some scientific ‘diplomats’ have called for a ‘proof of principle’, asking for a demonstration that protein pattern profiling is robust and reproducible, while the question regarding the identity of the peaks could be addressed at a later time point (29).
Since the Food and Drug Administration (FDA) made biomarkers part of the ‘critical path’ initiative in early 2004, efforts are under way to set up internal working groups as well as to foster collaborations between the FDA, the National Cancer Institute (NCI), industry and academia. Historically there has been little focus on biomarker discovery, but during the past few years several major companies (i.e. Roche, Novartis, Glaxo-SmithKline, Merck) have invested in biomarker discovery programs.
In addition to early detection and monitoring of disease processes, biomarkers have the potential to reduce the duration and thus the costs of clinical development efforts by gauging a drug’s effectiveness. Drugs are typically approved on the basis of clinical endpoints, namely on the basis of their manifest effects on the clinical symptoms of a disease, such as reduced tremor for anticholinergics in Parkinson’s patients, or fewer seizures for anticonvulsants. However, in some cases the proposed clinical benefit of a cancer drug on survival, for example, might not be detectable in clinical trials of reasonable duration. If biomarkers could be tied to a clinical outcome then they could be used as surrogate endpoints in such trials. These considerations became critical during the search for effective treatments for the life threatening disease Acquired Immune Deficiency Syndrome (AIDS), and in 1992, according to the Accelerated Approval provisions, the FDA approved a drug based on a surrogate marker and not a clinical outcome for the first time. Nowadays the FDA is leaning toward granting accelerated approvals for oncology drugs in much the same way.
It is a double-edged sword; there is urgency in getting life-saving drugs to the market, but on the other hand caution is advised. Once pharmaceutical companies have accelerated approval based on a surrogate, the incentive to seek full approval with phase III trials proving drug efficacy with hard outcomes diminishes. Accelerated approval does not seem to be the pressing issue. Rather, should we not be more concerned with the discovery of inexpensive, noninvasive, sensitive and specific biomarkers and surrogates as a first step? Over the past decade the FDA has only approved roughly one new diagnostic marker per year, the reason for which may be the heterogeneity of the disease and/or the patients.
Conclusion
The proteomics-based approach delivers a powerful set of tools for the challenges of the post-genome era. The time for biomarker exploration is now, given the apparent shift in philosophy of the FDA and the availability of many new methodologies and technologies. More coordinated blood plasma biomarker discovery efforts, such as the HUPO PPP, are needed to enable us to probe the depths of the plasma pool. Perhaps, if we are able to descend far enough, we will be rewarded with more “raptures of the deep” in the form of candidate biomarkers with clinical value.




Acknowledgement
Work at the Center for Experimental Bioinformatics was supported by a grant from the Danish National Research Foundation.
References
- Austin GE, Mullins RH,Morin LG. Non-enzymic glycation of individual plasma proteins in normoglycemic and hyperglycemic patients. Clin Chem 1987; 33: 2220-2224.
- Schenk S, Schoenhals GJ,Mann M. A plasma protein reference set identified with extremely high confidence through two consecutive stages of tandem mass spectrometry and ppm precursor mass accuracy. 2006; In preparation.
- Omenn GS. Special Issue: EXPLORING THE HUMAN PLASMA PROTEOME The HUPO Plasma Proteome Project (HPPP). Journal 2005; 5 (13): 3223-3549.
- Omenn GS, States DJ, Adamski M, Blackwell TW, Menon R, Hermjakob H, Apweiler R, Haab BB, Simpson RJ, Eddes JS, Kapp EA, Moritz RL, Chan DW, Rai AJ, Admon A, Aebersold R, Eng J, Hancock WS, Hefta SA, Meyer H, Paik YK, Yoo JS, Ping P, Pounds J, Adkins J, Qian X, Wang R, Wasinger V, Wu CY, Zhao X, Zeng R, Archakov A, Tsugita A, Beer I, Pandey A, Pisano M, Andrews P, Tammen H, Speicher DW,Hanash SM. Overview of the HUPO Plasma Proteome Project: results from the pilot phase with 35 collaborating laboratories and multiple analytical groups, generating a core dataset of 3020 proteins and a publicly-available database. Proteomics 2005; 5: 3226-3245.
- Adkins JN, Monroe ME, Auberry KJ, Shen Y, Jacobs JM, Camp DG, 2nd, Vitzthum F, Rodland KD, Zangar RC, Smith RD,Pounds JG. A proteomic study of the HUPO Plasma Proteome Project’s pilot samples using an accurate mass and time tag strategy. Proteomics 2005; 5: 3454-3466.
- Tammen H, Schulte I, Hess R, Menzel C, Kellmann M, Mohring T,Schulz-Knappe P. Peptidomic analysis of human blood specimens: comparison between plasma specimens and serum by differential peptide display. Proteomics 2005; 5: 3414-3422.
- He P, He HZ, Dai J, Wang Y, Sheng QH, Zhou LP, Zhang ZS, Sun YL, Liu F, Wang K, Zhang JS, Wang HX, Song ZM, Zhang HR, Zeng R,Zhao X. The human plasma proteome: analysis of Chinese serum using shotgun strategy. Proteomics 2005; 5: 3442-3453.
- Anderson NL, Polanski M, Pieper R, Gatlin T, Tirumalai RS, Conrads TP, Veenstra TD, Adkins JN, Pounds JG, Fagan R,Lobley A. The human plasma proteome: a nonredundant list developed by combination of four separate sources. Mol Cell Proteomics 2004; 3: 311-326.
- Blomberg A, Blomberg L, Norbeck J, Fey SJ, Larsen PM, Larsen M, Roepstorff P, Degand H, Boutry M, Posch A,et al. Interlaboratory reproducibility of yeast protein patterns analyzed by immobilized pH gradient two-dimensional gel electrophoresis. Electrophoresis 1995; 16: 1935-1945.
- Molloy MP, Brzezinski EE, Hang J, McDowell MT,VanBogelen RA. Overcoming technical variation and biological variation in quantitative proteomics. Proteomics 2003; 3: 1912-1919.
- Rai AJ, Gelfand CA, Haywood BC, Warunek DJ, Yi J, Schuchard MD, Mehigh RJ, Cockrill SL, Scott GB, Tammen H, Schulz-Knappe P, Speicher DW, Vitzthum F, Haab BB, Siest G,Chan DW. HUPO Plasma Proteome Project specimen collection and handling: towards the standardization of parameters for plasma proteome samples. Proteomics 2005; 5: 3262-3277.
- Lee HJ, Lee EY, Kwon MS,Paik YK. Biomarker discovery from the plasma proteome using multidimensional fractionation proteomics. Curr Opin Chem Biol 2006; 10: 42-49.
- Zhou M, Lucas DA, Chan KC, Issaq HJ, Petricoin EF, 3rd, Liotta LA, Veenstra TD,Conrads TP. An investigation into the human serum “interactome”. Electrophoresis 2004; 25: 1289-1298.
- Zhang X, Leung SM, Morris CR,Shigenaga MK. Evaluation of a novel, integrated approach using functionalized magnetic beads, bench-top MALDI-TOF-MS with prestructured sample supports, and pattern recognition software for profiling potential biomarkers in human plasma. J Biomol Tech 2004; 15: 167-175.
- Tanaka K, Waki H, Ido Y, Akita S, Yoshida Y,Yoshida T. Protein and Polymer Anaylsis up to m/z 100000 by Laser Ionization Time-of-Flight Mass Spectrometry. Rapid Comm. Mass Spectrom. 1988; 2: 151-158.
- Whitehouse CM, Dreyer RN, Yamashita M,Fenn JB. Electrospray interface for liquid chromatographs and mass spectrometers. Anal Chem 1985; 57: 675-679.
- Fenn JB, Mann M, Meng CK, Wong SF,Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science 1989; 246: 64-71.
- Olsen JV, de Godoy LM, Li G, Macek B, Mortensen P, Pesch R, Makarov A, Lange O, Horning S,Mann M. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol Cell Proteomics 2005; 4: 2010-2021.
- Semmes OJ, Feng Z, Adam BL, Banez LL, Bigbee WL, Campos D, Cazares LH, Chan DW, Grizzle WE, Izbicka E, Kagan J, Malik G, McLerran D, Moul JW, Partin A, Prasanna P, Rosenzweig J, Sokoll LJ, Srivastava S, Srivastava S, Thompson I, Welsh MJ, White N, Winget M, Yasui Y, Zhang Z,Zhu L. Evaluation of serum protein profiling by surface-enhanced laser desorption/ionization time-of-flight mass spectrometry for the detection of prostate cancer: I. Assessment of platform reproducibility. Clin Chem 2005; 51: 102-112.
- Wilson AM, Ryan MC,Boyle AJ. The novel role of C-reactive protein in cardiovascular disease: risk marker or pathogen. Int J Cardiol 2006; 106: 291-297.
- Anand IS, Latini R, Florea VG, Kuskowski MA, Rector T, Masson S, Signorini S, Mocarelli P, Hester A, Glazer R,Cohn JN. C-reactive protein in heart failure: prognostic value and the effect of valsartan. Circulation 2005; 112: 1428-1434.
- Palmerini T, Marzocchi A, Marrozzini C, Ortolani P, Saia F, Bacchi-Reggiani L, Virzi S, Gianstefani S,Branzi A. Preprocedural levels of C-reactive protein and leukocyte counts predict 9-month mortality after coronary angioplasty for the treatment of unprotected left main coronary artery stenosis. Circulation 2005; 112: 2332-2338.
- Wagner PD, Verma M,Srivastava S. Challenges for biomarkers in cancer detection. Ann N Y Acad Sci 2004; 1022: 9-16.
- Petricoin EF, Ardekani AM, Hitt BA, Levine PJ, Fusaro VA, Steinberg SM, Mills GB, Simone C, Fishman DA, Kohn EC,Liotta LA. Use of proteomic patterns in serum to identify ovarian cancer. Lancet 2002; 359: 572-577.
- Diamandis EP. Analysis of serum proteomic patterns for early cancer diagnosis: drawing attention to potential problems. J Natl Cancer Inst 2004; 96: 353-356.
- Diamandis EP. Mass spectrometry as a diagnostic and a cancer biomarker discovery tool: opportunities and potential limitations. Mol Cell Proteomics 2004; 3: 367-378.
- Diamandis EP. Point: Proteomic patterns in biological fluids: do they represent the future of cancer diagnostics? Clin Chem 2003; 49: 1272-1275.
- Baggerly KA, Morris JS,Coombes KR. Reproducibility of SELDI-TOF protein patterns in serum: comparing datasets from different experiments. Bioinformatics 2004; 20: 777-785.
- Ransohoff DF. Evaluating discovery-based research: when biologic reasoning cannot work. Gastroenterology 2004; 127: 1028.
- Adkins JN, Varnum SM, Auberry KJ, Moore RJ, Angell NH, Smith RD, Springer DL,Pounds JG. Toward a human blood serum proteome: analysis by multidimensional separation coupled with mass spectrometry. Mol Cell Proteomics 2002; 1: 947-955.
- Pieper R, Gatlin CL, Makusky AJ, Russo PS, Schatz CR, Miller SS, Su Q, McGrath AM, Estock MA, Parmar PP, Zhao M, Huang ST, Zhou J, Wang F, Esquer-Blasco R, Anderson NL, Taylor J,Steiner S. The human serum proteome: display of nearly 3700 chromatographically separated protein spots on two-dimensional electrophoresis gels and identification of 325 distinct proteins. Proteomics 2003; 3: 1345-1364.
- Chan KC, Lucas DA, Hise D, Schaefer CF, Xiao Z, Janini GM, Buetow KH, Issaq HJ, Veenstra TD,Conrads TP. Analysis of the Human Serum Proteome. Clin Prot J 2004; 1: 101-226.
- Shen Y, Jacobs JM, Camp DG, 2nd, Fang R, Moore RJ, Smith RD, Xiao W, Davis RW,Tompkins RG. Ultra-high-efficiency strong cation exchange LC/RPLC/MS/MS for high dynamic range characterization of the human plasma proteome. Anal Chem 2004; 76: 1134-1144.
- Rose K, Bougueleret L, Baussant T, Bohm G, Botti P, Colinge J, Cusin I, Gaertner H, Gleizes A, Heller M, Jimenez S, Johnson A, Kussmann M, Menin L, Menzel C, Ranno F, Rodriguez-Tome P, Rogers J, Saudrais C, Villain M, Wetmore D, Bairoch A,Hochstrasser D. Industrial-scale proteomics: from liters of plasma to chemically synthesized proteins. Proteomics 2004; 4: 2125-2150.
- Jin WH, Dai J, Li SJ, Xia QC, Zou HF,Zeng R. Human plasma proteome analysis by multidimensional chromatography prefractionation and linear ion trap mass spectrometry identification. J Proteome Res 2005; 4: 613-619.
- Emerson RE,Ulbright TM. The use of immunohistochemistry in the differential diagnosis of tumors of the testis and paratestis. Semin Diagn Pathol 2005; 22: 33-50.
- Johnson PJ. The role of serum alpha-fetoprotein estimation in the diagnosis and management of hepatocellular carcinoma. Clin Liver Dis 2001; 5: 145-159.
- Marrero JA, Su GL, Wei W, Emick D, Conjeevaram HS, Fontana RJ,Lok AS. Des-gamma carboxyprothrombin can differentiate hepatocellular carcinoma from nonmalignant chronic liver disease in american patients. Hepatology 2003; 37: 1114-1121.
- Verma S, Szmitko PE,Ridker PM. C-reactive protein comes of age. Nat Clin Pract Cardiovasc Med 2005; 2: 29-36; quiz 58.
- Vermeire S, Van Assche G,Rutgeerts P. Laboratory markers in IBD: useful, magic, or unnecessary toys? Gut 2006; 55: 426-431.
- Ndrepepa G, Kastrati A, Braun S, Mehilli J, Niemoller K, von Beckerath N, von Beckerath O, Vogt W,Schomig A. N-terminal probrain natriuretic peptide and C-reactive protein in stable coronary heart disease. Am J Med 2006; 119: 355 e351-358.
- Duffy MJ. Clinical uses of tumor markers: a critical review. Crit Rev Clin Lab Sci 2001; 38: 225-262.
- Rosenthal AN,Jacobs IJ. The role of CA 125 in screening for ovarian cancer. Int J Biol Markers 1998; 13: 216-220.
- Xu Y, Shen Z, Wiper DW, Wu M, Morton RE, Elson P, Kennedy AW, Belinson J, Markman M,Casey G. Lysophosphatidic acid as a potential biomarker for ovarian and other gynecologic cancers. Jama 1998; 280: 719-723.
- Verma M,Srivastava S. New cancer biomarkers deriving from NCI early detection research. Recent Results Cancer Res 2003; 163: 72-84; discussion 264-266.
- Bast RC, Jr., Badgwell D, Lu Z, Marquez R, Rosen D, Liu J, Baggerly KA, Atkinson EN, Skates S, Zhang Z, Lokshin A, Menon U, Jacobs I,Lu K. New tumor markers: CA125 and beyond. Int J Gynecol Cancer 2005; 15 Suppl 3: 274-281.
- Fields MM,Chevlen E. Ovarian cancer screening: a look at the evidence. Clin J Oncol Nurs 2006; 10: 77-81.
- Lloyd KO, Yin BW,Kudryashov V. Isolation and characterization of ovarian cancer antigen CA 125 using a new monoclonal antibody (VK-8): identification as a mucin-type molecule. Int J Cancer 1997; 71: 842-850.
- Posner MR,Mayer RJ. The use of serologic tumor markers in gastrointestinal malignancies. Hematol Oncol Clin North Am 1994; 8: 533-553.
- Macdonald JS. Carcinoembryonic antigen screening: pros and cons. Semin Oncol 1999; 26: 556-560.
- Zhao Y, Verselis SJ, Klar N, Sadowsky NL, Kaelin CM, Smith B, Foretova L,Li FP. Nipple fluid carcinoembryonic antigen and prostate-specific antigen in cancer-bearing and tumor-free breasts. J Clin Oncol 2001; 19: 1462-1467.
- Goldstein MJ,Mitchell EP. Carcinoembryonic antigen in the staging and follow-up of patients with colorectal cancer. Cancer Invest 2005; 23: 338-351.
- Duffy MJ. Serum tumor markers in breast cancer: are they of clinical value? Clin Chem 2006; 52: 345-351.
- Liang X, Smoller BR,Golitz LE. Expression of CD44 and CD44v6 in primary cutaneous CD30 positive T-cell lymphoproliferative disorders. J Cutan Pathol 2002; 29: 459-464.
- Akisik E, Bavbek S,Dalay N. CD44 variant exons in leukemia and lymphoma. Pathol Oncol Res 2002; 8: 36-40.
- Lossos IS,Morgensztern D. Prognostic biomarkers in diffuse large B-cell lymphoma. J Clin Oncol 2006; 24: 995-1007.
- Garcea G, Neal CP, Pattenden CJ, Steward WP,Berry DP. Molecular prognostic markers in pancreatic cancer: a systematic review. Eur J Cancer 2005; 41: 2213-2236.
- Endo K,Terada T. Protein expression of CD44 (standard and variant isoforms) in hepatocellular carcinoma: relationships with tumor grade, clinicopathologic parameters, p53 expression, and patient survival. J Hepatol 2000; 32: 78-84.
- Wang CS, Lin CL, Lee HC, Chen KY, Chiang MF, Chen HS, Lin TJ,Liao LY. Usefulness of serum des-gamma-carboxy prothrombin in detection of hepatocellular carcinoma. World J Gastroenterol 2005; 11: 6115-6119.
- Pegram M,Slamon D. Biological rationale for HER2/neu (c-erbB2) as a target for monoclonal antibody therapy. Semin Oncol 2000; 27: 13-19.
- Kuerer HM, Thompson PA, Krishnamurthy S, Fritsche HA, Marcy SM, Babiera GV, Singletary SE, Cristofanilli M, Sneige N,Hunt KK. High and differential expression of HER-2/neu extracellular domain in bilateral ductal fluids from women with unilateral invasive breast cancer. Clin Cancer Res 2003; 9: 601-605.
- Sternlicht MD. Key stages in mammary gland development: the cues that regulate ductal branching morphogenesis. Breast Cancer Res 2006; 8: 201.
- Mehra R, Huria A, Gupta P,Mohan H. Choriocarcinoma with negative urinary and serum beta human chorionic gonadotropin (betaHCG)–a case report. Indian J Med Sci 2005; 59: 538-541.
- Stenman UH, Alfthan H,Hotakainen K. Human chorionic gonadotropin in cancer. Clin Biochem 2004; 37: 549-561.
- O’Brien N, Maguire TM, O’Donovan N, Lynch N, Hill AD, McDermott E, O’Higgins N,Duffy MJ. Mammaglobin a: a promising marker for breast cancer. Clin Chem 2002; 48: 1362-1364.
- Fleming TP,Watson MA. Mammaglobin, a breast-specific gene, and its utility as a marker for breast cancer. Ann N Y Acad Sci 2000; 923: 78-89.
- Bernstein JL, Godbold JH, Raptis G, Watson MA, Levinson B, Aaronson SA,Fleming TP. Identification of mammaglobin as a novel serum marker for breast cancer. Clin Cancer Res 2005; 11: 6528-6535.
- Kim JH, Skates SJ, Uede T, Wong KK, Schorge JO, Feltmate CM, Berkowitz RS, Cramer DW,Mok SC. Osteopontin as a potential diagnostic biomarker for ovarian cancer. Jama 2002; 287: 1671-1679.
- Zheng Y, Xu Y, Ye B, Lei J, Weinstein MH, O’Leary MP, Richie JP, Mok SC,Liu BC. Prostate carcinoma tissue proteomics for biomarker discovery. Cancer 2003; 98: 2576-2582.
- Mok SC, Chao J, Skates S, Wong K, Yiu GK, Muto MG, Berkowitz RS,Cramer DW. Prostasin, a potential serum marker for ovarian cancer: identification through microarray technology. J Natl Cancer Inst 2001; 93: 1458-1464.
- Mills GB, Bast RC, Jr.,Srivastava S. Future for ovarian cancer screening: novel markers from emerging technologies of transcriptional profiling and proteomics. J Natl Cancer Inst 2001; 93: 1437-1439.
- Lilja H, Oldbring J, Rannevik G,Laurell CB. Seminal vesicle-secreted proteins and their reactions during gelation and liquefaction of human semen. J Clin Invest 1987; 80: 281-285.
- Babuin L,Jaffe AS. Troponin: the biomarker of choice for the detection of cardiac injury. Cmaj 2005; 173: 1191-1202.
- Collinson PO, Gaze DC, Stubbs PJ, Swinburn J, Khan M, Senior R,Lahiri A. Diagnostic and prognostic role of cardiac troponin I (cTnI) measured on the DPC Immulite. Clin Biochem 2006;
- Nagahara D, Nakata T, Hashimoto A, Takahashi T, Kyuma M, Hase M, Tsuchihashi K,Shimamoto K. Early positive biomarker in relation to myocardial necrosis and impaired fatty acid metabolism in patients presenting with acute chest pain at an emergency room. Circ J 2006; 70: 419-425.




