Setting new standards in Ultra HTS
Posted: 2 February 2006 | | No comments yet
Ultra high-throughput screening (UHTS) offers the possibility to discover novel pharmacophores. To benefit from UHTS special demands concerning assay quality and data analysis have to be met.
Ultra high-throughput screening (UHTS) offers the possibility to discover novel pharmacophores. To benefit from UHTS special demands concerning assay quality and data analysis have to be met.
Ultra high-throughput screening (UHTS) offers the possibility to discover novel pharmacophores. To benefit from UHTS special demands concerning assay quality and data analysis have to be met.
Drug discovery is one of the most challenging missions of the investigative pharmaceutical industry. The need for novel lead structures forced the companies to build up big compound libraries containing often more than a million different substances. Bottlenecks such as compound management and compound screening had to be alleviated by automation to make use of these growing libraries.
Full automation and miniaturisation of the screening process allows Bayer to screen more than 1.7 million single compounds in eight working days against a target. A range of target classes with a high relevance to drug discovery are screened: GPCRs, ion channels, transporters, nuclear receptors, proteases, kinases, phosphatases, phosphodiesterases, polymerases and others. Screening robots with a throughput of more than 250,000 test points per day and the miniaturisation of all assays to the 1536-well format allow for ultra high-throughput screening (UHTS). The high degree of miniaturisation poses high demands on the assays performed. Robustness and stability have to be combined with high sensitivity. This review will give insights into the lead finding process at Bayer HealthCare describing assay principles and data analysis.
Cell based assay systems
Presently cell-based assays are becoming more important in the area of HTS and even UHTS, because they offer the relevant biological background to perform screening under physiological conditions. Issues such as toxicity and membrane permeability can be addressed already during the primary screen of cellular assays. On the other hand, cell based assays are characterised by their increased complexity. Assay development can be more resource and time consuming compared to enzymatic or binding assays, e.g. the generation of cell lines. To minimise resources and time, several steps of the cell generation process at Bayer have been automated. Upon transfection with the gene of interest cells are seeded on 1536-well microtiter plates (MTPs). After formation of single colonies, more than 10.000 stably transfected clones can be tested under assays conditions, which are similar to that of the intended UHTS. The best clones are selected and subcloned under the same conditions. Sensitivity, signal intensity and growth rate are the main criteria for clone selection. This clone selection process allows rapid and efficient generation of cell lines suitable for UHTS.
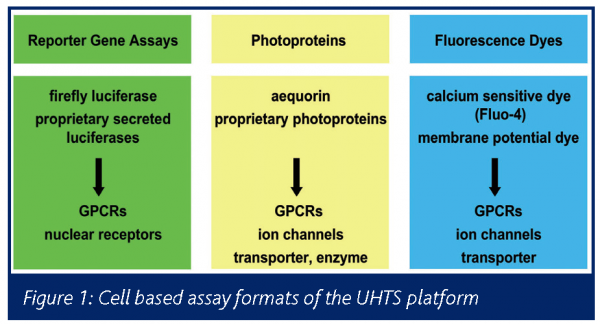
Readout technologies for cell-based assays have to be adapted to the demands of UHTS in terms of robustness and stability. This means low standard deviations on the 1536-well MTPs and constant sensitivity and signal intensity during 20 hour screening runs. The assay technologies adapted to the Bayer UHTS platform include luciferase and aequorin1 based luminescent readouts and fluorescent readouts using calcium indicators and membrane potential dyes (Figure1).
Reporter gene assays with firefly luciferase or a proprietary secreted luciferase are performed for screening of Gs or Gi coupled GPCRs and nuclear receptors. Calcium transients induced by receptor or ion channel stimulation are monitored with aequorin or Fluo-4. Activity of ion channels and electrogenic transporter is detected by the use of membrane potential dyes.
For most targets different assay formats are developed, to identify the most suitable technique. The use of an assay with alternative readout for the same target in secondary screening allows early identification of pathway dependent false positives and assay artifacts.
High hit rates due to unspecific and toxic effects of compounds are a major problem of cell-based assays. Elimination of these compounds is essential for screening of comprehensive libraries and adequate strategies have to be integrated into the screening process.
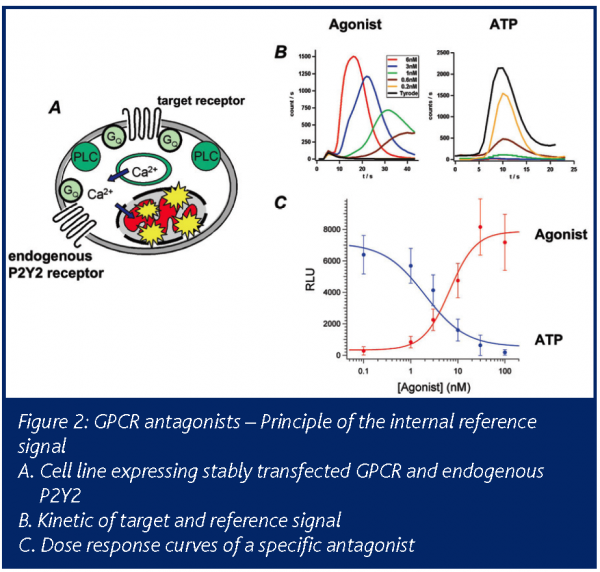
For GPCR antagonist screens with aequorin readout we make use of the endogenously expressed P2Y2 receptor. Specific antagonists as well as toxic compounds reduce the signal of the transfected target receptor and cannot be distinguished from each other. But activation of the endogenous P2Y2 receptor with ATP subsequent to target stimulation allows differentiation of antagonists from unspecific or toxic compounds, because the receptors are coupled to the same signal cascade. In Figure 2 the principle of the internal reference signal is depicted. Activation of the target receptor by a specific agonist leads to reduction of the P2Y2 response due to pathway desensitisation and aequorin consumption. Blocking of the pathway by a target receptor antagonists allows activation of P2Y2, while toxic compounds inhibit its activity. By its action the endogenous receptor mirrors the activity of the target receptor and can be used as a highly relevant internal control.
For inhibitory reporter gene assays the combination of firefly luciferase and a secreted luciferase2 allows subsequent detection of target and reference signal, respectively. The firefly signal is coupled to activation of the target, while the secreted luciferase is expressed constitutively. Mutual decrease of both signals indicates an unspecific effect of a compound. The internal reference signals described here allow efficient elimination of unspecific compounds.
Cell based assays offer the advantages of physiological screening conditions and high information content. A high degree of automation and versatile solutions for early hit validation allowed Bayer to adopt cell based assays to its UHTS platform.
Biochemical assay systems
In early drug discovery, enzymes make up a substantial part of the drug targets of major interest. In biochemical assay systems enzyme modulators (inhibitors or activators) are investigated in a simple experimental setting where parameters such as membrane permeability or serum binding do not complicate the measurement of the modulator-enzyme interaction. Nevertheless, sophisticated assay systems for UHTS in 1536-well MTPs have been developed by Bayer scientists, which include complex pipetting schemes and multiple measurements to identify novel leads that bind and thereby specifically modulate the enzymes function.
For numerous target classes we have implemented unique assay systems to identify enzyme inhibitors. The enzyme activity is monitored in a functional assay where the signal (most frequently fluorescence or luminescence) is either directly generated by the action of the enzyme or indirectly via a cascade of enzymes facilitating the measurement of otherwise difficult to detect enzymatic reaction products or substrates. Changes of the signal indicate the action of an enzyme modulator.
A complex but well established enzyme cascade to measure the activity of kinases with the aim to identify specific kinase inhibitors by UHTS will be exemplified and discussed here.
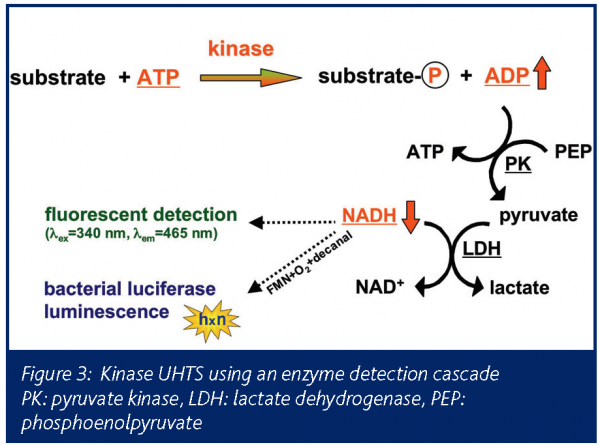
This system (Figure 3) meets the challenge to develop a sensitive, broadly applicable and robust kinase assay system with no principal restriction with respect to the substrate choice and the ability to use µM to mM ATP concentrations: The formation of ADP from ATP during the kinase catalysed reaction is coupled to the oxidation of NADH to NAD+ via pyruvate kinase and lactate dehydrogenase3. NADH consumption can be measured as the kinase reaction proceeds.
The detection of ADP makes the system applicable to any protein as well as non-protein kinase or even ATPase. Furthermore, the use of label-free kinase substrates greatly facilitates studies on the kinases’ substrate specificity by screening peptide libraries. Combined with the possibility of using a wide range of ATP concentrations it enables us to establish multiple assay conditions for identifying new leads.
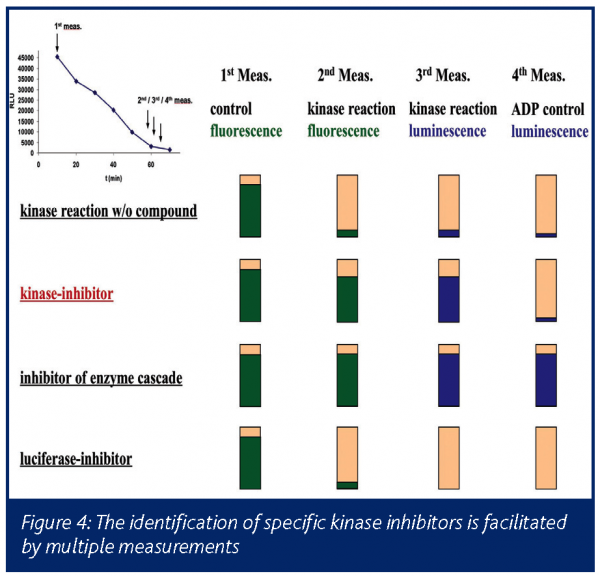
The final signal of the enzyme cascade can be either NADH fluorescence (continuous measurement possible) or the directly derived bacterial luciferase luminescence. In many cases at Bayer both measurements were employed subsequently during UHTS to be able to identify kinase inhibitors independent of their interference with either of the two principles of measurement. Typically additional reference measurements are included: A first reference measurement prior to the kinase reaction to identify autofluorescent compounds and a second post-kinase reaction measurement upon addition of an ADP excess to distinguish cascade inhibitors from true kinase inhibitors. Complex pipetting schemes have been implemented in order to realise control reactions and measurements (Figure 4).
In this context it is interesting to note that multiple measurements are not time critical in Bayer’s UHTS due to fluorescence and luminescence imaging technology reading a 1536-well MTP in less than 30 s.
Despite the obvious complexity of the system, in our hands it proved to be sensitive, reliable, robust and highly reproducible. Thus we favour it for our 1536-well UHTS over other available kinase assay systems. The additional control reactions and measurements have proven to increase data quality significantly as they allow us to distinguish unspecific compound effects on cascade enzymes and measurement artifacts from specific kinase inhibition.
Information management and data analysis
During the past few years, the introduction of UHTS technologies, new assay design and detection technologies has exponentially increased the amount and complexity of screening data. While today many commercial software packages have been built for tracking, handling and viewing measured assay activity data, including corresponding chemical data, ten years ago no adequate software was available. Therefore at Bayer Healthcare a tailored Laboratory Information Management System (LIMS) was developed and continuously improved in parallel with the introduction of laboratory automation to cope with the pharmacological data generated by the innovative cell based and biochemical assays. This in-house system is used to perform hit identification as well as controlling of the robotic system for hit picking and compound formatting for confirmative hit validation.
Besides flexibility to integrate multiple pharmacological measurements, as well as multiple reference measurements from various spectrometers, computational runtime performance for data processing of routinely tested – often more than 1.7 million – compounds was one of the most important requirements for this software. It allows for standardised quality control of every day assay results and efficient identification of experimental problems and artifacts. Data processing includes normalisation of measured signals to ensure comparability of results of different robotic test days with possibly varying sensitivity and signal to background noise.
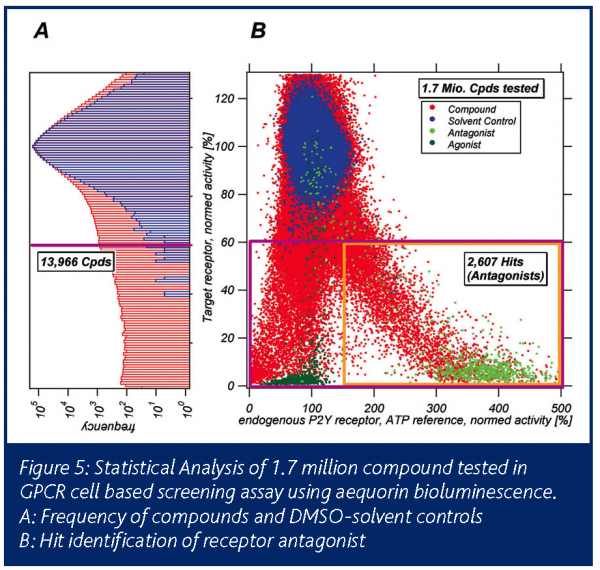
Figure 5 visualises the analysis of UHTS data for a cell based GPCR assay. Scatter plot (A): on the y-axis the signals of the target receptor are plotted, on the x-axis the signals of the endogenous P2Y2 reference receptor. The activity values for all individual 1.7 million compounds are shown in blue. All activity values were normalised to the median value of solvent reference (DMSO) available on every MTP. Individual variability of these solvent controls is shown in blue dots. The effect of the natural peptide ligand in various concentrations is shown in dark green. Values of a target reference antagonist are shown in light green.
The accumulated frequency for the DMSO controls is shown in the histogram (B): the standard deviation is below 7.8 per cent. The hit limit was determined by the compound to DMSO control ratio for the activity of the target receptor: less than 1 per cent of the measured DMSO samples should be observed at the hit limit cutoff. This permits an acceptable low rate of statistical errors. Use of the transgenic target receptor signal only would result in 13,988 primary hits. Including the reference signal of the endogenous receptor allows discrimination of receptor antagonists from unspecific compounds and results in 2,607 hits for confirmatory screening.
A very important step is the detection and correction of systematic errors e.g. errors in liquid handling; decay of cells; positional effects of detectors; reagent evaporation or compound cross contamination. Such errors can cause either overestimation (false positive hits) or, even more severe, underestimation (false negative hits) of measured compound effect data4. After the primary screening a hit picking process is carried out in 1536-well MTPs to allow for efficient exclusion of remaining false positive hits in confirmatory UHTS. Confirmatory screening is performed in multiple concentrations using the primary screening assay as well as reference assays, which can be assays with alternative readouts or different target assays, to evaluate selectivity and specificity of the hits. In order to get hints on false negative compounds, chemoinformatic tools are applied to include structurally similar compounds in confirmatory UHTS. Chemoinformatic techniques5 such as clustering, similarity searching and initial pharmacophore identification assist in understanding relationships between biological data and properties of compounds. These tools allow generating initial structure-activity-relationships from confirmatory UHTS data.
In summary, UHTS is a powerful approach for the discovery of novel pharmacophors. Screening success depends on the combination of versatile assay technologies, automation and data analysis. Based on a ten year experience and constant enhancements, Bayer Healthcare developed a UHTS platform, which is now an essential part of the drug discovery process.





References
- Dupriez VJ, Maes K, Le Poul E, Burgeon E, Detheux M: Aequorin-based functional assays for G-protein-coupled receptors, ion channels, and tyrosine kinase receptors. Receptors Channels. 2002; 8 (5-6): 319-330.
- Markova SV, Golz S, Frank LA, Kalthof B, Vysotski ES: Cloning and expression of cDNA for a luciferase from the marine copepod Metridia longa. A novel secreted bioluminescent reporter enzyme. J Biol Chem. 2004 Jan 30; 279 (5): 3212-3217
- Williamson JR, Corkey BE: Adenosine 5’-diphosphate and adenosine monophosphate determination with pyruvate kinase, myokinase, and lactate dehydrogenase. Methods Enzymol. 1969; 13: 494-497.
- Brideau C, Gunter B, Pikounis B, Liaw A: Improved Statistical Methods for Hit Selection in High-Throughput Screening. Journal of Biomolecular Screening, 2003, 8 (6) 634- 647
- Olsson T, Oprea T.A.: Cheminformatics: A tool for decision-makers in drug discovery. Current Opin. Drug Discov. Dev. 2001, 4 (3), 308-313




