Microbial detection in mammalian cell culture systems
Posted: 11 November 2005 | | No comments yet
Timely results for microbial bioburden monitoring of in-process cell culture samples are critical to the production process for recombinant proteins and other biopharmaceutical products.
Timely results for microbial bioburden monitoring of in-process cell culture samples are critical to the production process for recombinant proteins and other biopharmaceutical products.
Timely results for microbial bioburden monitoring of in-process cell culture samples are critical to the production process for recombinant proteins and other biopharmaceutical products.
One type of cell culture manufacturing process, called a batch refeed process, can be used to demonstrate two main aspects of the cell culture operation which will further exemplify the importance of timely results. The first aspect is the nature of the refeed process itself. In a typical batch refeed process, 80-90% of the contents of a bioreactor are removed (harvested) every 3-4 days. The remaining 10-20% serves as the inoculum for the next bioreactor batch, created by adding fresh medium to the bioreactor and then repeating the entire sequence of operations at each interval of time. Owing to the time required for results using traditional bioburden monitoring assays, the cell culture process must continue at risk until the results are available. The contents that are harvested from the bioreactor are purified through numerous separation/purification steps to generate the bulk drug substance. The critical impact on timely results at this harvesting stage of the process is that multiple bioreactors are harvested at the same time and pooled together for purification. Operational decisions to refeed the bioreactor or pool the harvests are often made without the benefit of microbiological data. By the time results are available, one or more bioreactors could be lost to contamination.
Typical bioburden monitoring is performed using membrane filtration, followed by transfer of the membrane to a general-purpose agar and incubation of the agar plates. This growth-based method can produce results in a minimum 1-7 days, when the plates are examined for the macroscopic formation of colonies. Based on the retrospective nature of the availability of bioburden results (as stated above, the manufacturing process proceeds at risk during the incubation period of the agar plates), it can be seen how a rapid result can reduce the risk to the process and provide greater process control. It is important to note that the bioburden testing described here is an in-process test, not a release test. Furthermore, although a monoculture of mammalian cells is expected in the cell culture process, the presence of a certain level of bioburden can be tolerated with validated removal systems in place during downstream purification (sterility is not a requirement at this stage, where the presence of bioburden is an operational concern for the process).
In contrast to the growth-based bioburden assay, the ScanRDI™ system rapidly detects individual viable microorganisms in filterable solutions (without the need for the growth of the microorganisms into visible colonies). Results are available on the same day the sample is submitted, reducing process risk necessitated by the growth-based assays.
In order to evaluate the premise that the ScanRDI™ assay is comparable to the growth-based membrane filtration assay for the purpose of detecting the presence of bioburden in cell culture samples, a master validation plan was drafted at Wyeth Biotech, Andover MA (USA), including elements of method evaluation, concurrent testing, method validation and crossover studies. A summary of the master plan will be presented and the results of the concurrent testing will be described in detail.
Principle of the method
In the ScanRDI™ assay, samples are passed through a 0.45μ filter to retain microorganisms, which are then labeled with reagents to determine their viability. Only viable organisms are capable of enzymatically cleaving the non-fluorescent substrate and retaining the fluorescent end products. The entire surface of the membrane is then analysed by the ScanRDI™ laser scanning cytometer, which detects and counts any labeled cell on the filter. The detection and enumeration of microorganisms is achieved by the system through a sequential series of computerised algorithms, which discriminate between fluorescent signal generated by bacterial cells and nonspecific fluorescence generated by debris or other material on the filter membrane. Each labeled microorganism can then be visually observed using a fluorescent microscope.
In the application described herein, the ScanRDI™ assay is intended to detect a low-level bacterial contamination in a background of millions of Chinese Hamster Ovary (CHO) cells. This assay is intended to be a qualitative, limit type of assay, indicating the presence of bacteria above a control limit within a CHO cell culture. The assay is not designed to quantify the level of bacteria present, nor is it intended to be a sterility test.
Because the principle of detection of the ScanRDI™ system is based on the uptake and cleavage of a ubiquitous fluorescent viability indicator, the instrument will detect viable CHO cells in addition to any bacteria present. Intact CHO cells themselves would be too large to pass the discriminating algorithms for a bacterial signal, but they would contribute such a level of fluorescence that the system would not be able to distinguish any individual contaminating organisms from the highly fluorescent background. Therefore, a differential lysis protocol was employed per Standard Operating Procedures (SOPs) to preferentially eliminate the CHO cells while having minimal effect on any bacterial cells present within the culture.
The differential lysis procedure, while highly effective at reducing the signal generated by the mammalian cells, is not currently capable of completely eliminating CHO cell interference. Small pieces of CHO cell debris retained on the membrane surface can pass the discrimination of the software and be detected by the instrument as a bacterial or fungal cell, thus presenting the potential for interference with the detection of contamination. Therefore, there is a level below which a positive ScanRDI™ result is potentially inaccurate; this level is considered the baseline noise and was set as the control limit for a positive result for the assay.
The baseline noise for the assay was initially set during method development at 10 fluorescent events (fe)/mL (the term ’fluorescent events‘ has been assigned to the results generated by the ScanRDI™ for this application, rather than the term ’bacteria‘ or ’microorganism,’ because of the potential for this result to be caused by debris and not by a viable microorganism). This value of 10 fe/mL was used as the control limit during the initial stages of the validation; however, following the concurrent testing stage, the baseline noise was raised to 25 fe/mL. As mentioned above, the baseline noise was also set as the control limit for a positive result; therefore, for a result to be considered positive in this application, the ScanRDI™ must report a count of ≥25 fe/mL. Below this level, cell culture processing will continue; above this level, an alert will occur and an investigation will be initiated.
In order for the ScanRDI™ to be used as the bioburden monitoring tool further downstream in the manufacturing process, the baseline noise must decrease (as the specifications for bioburden are lower downstream in the process, less baseline noise can be tolerated in the assay) and as the baseline noise decreases (indicating less interference), the more appropriate term for the ScanRDI™ result would be ’bacteria‘ or ’viable microorganisms.’
Master plan overview
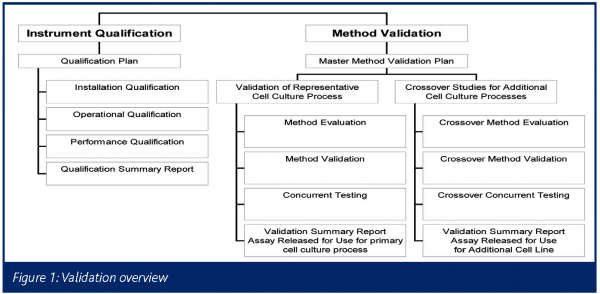
Figure 1 below provides an overview of the validation activities associated with the ScanRDI™. A summary of the method validation portion follows.
Method validation summary
The ScanRDI™ method for detecting bioburden in mammalian cell culture systems is being validated according to a master method validation plan. The validation consists of four phases: method evaluation, concurrent testing, method validation and crossover studies.
The general philosophy of the method validation was to use a representative CHO cell culture process for the initial validation testing, followed by a less extensive validation using crossover studies for each additional cell culture process. The concept of using a representative cell culture is supported because, while each process has undergone slight modifications, all processes were ultimately derived from closely related host cells (CHO). Therefore, any differences between cell culture processes are not expected to impact the basis of the ScanRDI™ assay.
To summarise the testing performed using the representative cell culture process, the method validation paradigm was to design a concurrent testing study (Concurrent Testing in Figure 1) to demonstrate that the ScanRDI™ was equivalent to, or better than, the growth-based membrane filtration in terms of qualitative detection of contamination in a timely manner. Due to the fact that this concurrent testing was designed to be performed using commercial in-process samples from the manufacturing facility, there was a high probability that the study would run to completion without any occurrence of contamination (which would result in both methods having all zero results, making it impossible to draw any statistical conclusions). Therefore, an additional study was designed to confirm the ability of the method to detect bacteria that were known to be present via spiking cell culture spinners (termed Method Evaluation in Figure 1). Finally, to complete the validation in accordance with current validation guidelines and established industry standards (ICH 1995a; ICH 1995b; USP <1225>; PDA 2000), a Method Validation Protocol was drafted.
After completion of the validation using the representative cell culture process, the proposed philosophy for additional processes is to execute the individual elements (concurrent testing, method evaluation and method validation) of the testing performed using the primary cell culture process, although with a narrower range of microorganisms in the case of method evaluation and method validation, or with a shorter period of concurrent testing (termed Crossover Studies in Figure 1). Upon completion of the validation activities for commercial products, a regulatory filing change will be required to replace the traditional method with the rapid ScanRDI™ method.
Concurrent testing
The intent of the concurrent testing was to demonstrate that under actual conditions of use for manufacturing samples in the QC environment, the ScanRDI™ would detect contamination at a rate equivalent to the growth-based membrane filtration method, with the additional benefit of providing results on the same day of testing.
The concurrent testing included spinners and bioreactors from the manufacturing facility, tested simultaneously using the ScanRDI™ and growth-based membrane filtration methods. At least 90% of the results from the two methodologies (ScanRDI™ and growth-based membrane filtration) were required to ’match‘ in order to meet the acceptance criteria. A ’matching‘ result was achieved when both methodologies agreed – both either positive or negative. A positive result on the ScanRDI™ was defined as ≥10 fe and a positive result on the membrane filtration was defined as ≥5 CFU (in order to avoid any false positives due to sample handling). Furthermore, in order to justify use in the routine operation, at least 90% of the samples tested on the ScanRDI™ were required to generate a result (there could be no more than 10% failures to obtain a ScanRDI™ result for any reason).
During the concurrent testing, 101 samples representing spinners, 250-L and 2500-L bioreactors were tested over 20 weeks. The results indicated qualitative equivalence in the rate of detection of contamination for the ScanRDI™ compared to the growth-based membrane filtration method. Furthermore, the ScanRDI™ gave no false negatives during concurrent testing (there were no instances where the growth-based membrane filtration was >5 CFU and the ScanRDI™ was <10 fe).
There were six instances of mismatches between the ScanRDI™ and growth-based membrane filtration; all were false positives for the ScanRDI™ (ScanRDI™ results > 10 fe with membrane filtration results of 0 CFU). Upon review of the data, all six false positives were < 25 fe on the ScanRDI™ (four results were less than 20 fe, while two results were 22 fe). The original control limit of 10 fe/mL on the ScanRDI™ was set based on assay development data. The data from the concurrent testing demonstrated that raising the limit for a positive result on the ScanRDI™ from 10 fe (based on development data using laboratory scale spinner cultures) to 25 fe (based on manufacturing samples from spinners and bioreactors) would reduce the current level of false positives to 0 compared to the growth-based membrane filtration and reduce the risk of future false positives. Even with a control limit of 25 fe, contamination would still be detected prior to growth on the membrane filtration plates (this is proven in the case study below, which describes a contamination event that occurred during concurrent testing). The increased level of false positives during the concurrent testing is proposed to be an effect of the robustness of the CHO cells under the more controlled bioreactor conditions (as opposed to laboratory scale spinners), leading to a potential for a slight reduction in the effectiveness of the lysis procedure.
The data generated during the concurrent testing demonstrated that setting the appropriate limit for a positive result is a balance of false positives versus false negatives. Setting it too low may require frequent investigations. Setting it too high increases the risk of a contamination being passed on to the next batch. With a control limit of 25, it is understood that if a contamination occurred at the level of 1-24 CFU with the growth-based assay, a potential false negative would occur using the ScanRDI™. In order to assess the impact of this potential, historical data for the growth-based bioburden assay was reviewed and assessed for the occurrence of true positives (contamination in the cell culture was confirmed) at the level of 1-24 CFU/mL. Furthermore, the rate of false positives for the growth-based membrane filtration assay was assessed (a false positive is defined as an initial positive result without confirming evidence of contamination in the cell culture, many times based on subsequent sampling information). Results of this analysis supported the premise that the ScanRDI™ with a control limit of 25fe/mL could provide equivalent control of bioburden in the cell culture process. In the instance of borderline results, there is the added benefit of the rapid ScanRDI™ assay having results for the suspect batch as well as results from the subsequent batch that either substantiate contamination or disprove it.
Example of ScanRDI™ in contamination detection
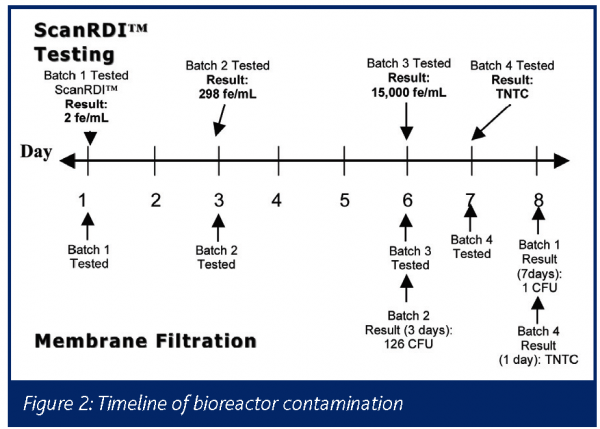
In the third month of the concurrent testing, the ScanRDI™ demonstrated its early detection capabilities when a bioreactor in the facility became contaminated. See Figure 2 for a timeline of the contamination. The ScanRDI™ first detected evidence of this contamination on Day 1, in Batch 1. However, the level in Batch 1 was below 10 fe/mL, so was not recorded as a positive result. The next batch (Batch 2) was tested two days later on Day 3. The ScanRDI™ result for that sample was 298 fe/mL. The first detection of bacteria with the traditional method – growth-based membrane filtration – was not until another five days later on Day 8 when the plate from Batch 1 was found to have 1 CFU/mL (the result of 1 CFU/mL, however, would imply a possible sample handling contamination rather than bioreactor contamination). The first definitive sign of contamination in the bioreactor from the growth-based membrane filtration was on Day 6, when a count of 126 CFU/mL was read on the plate from Batch 2. This result came three days later than the ScanRDI™ result for that same batch. If the ScanRDI™ assay had been validated for use prior to this contamination (even with a control limit of 25 fe/mL), the contamination would have been detected at least three days earlier than with the growth-based membrane filtration, leading to improved control of bioburden in the process and reducing the loss of bioreactor material due to the retrospective nature of the traditional results.
Conclusion
This article has described an ongoing validation effort at Wyeth BioPharma and the data has shown that even for a complex sample matrix such as mammalian cell culture, the ScanRDI™ can be validated. It can provide a greater level of control to the manufacturing process than the growth-based method due to the accelerated availability of results. The amount of time and effort to validate this technology is extensive and on-site knowledge of the system is essential for success. However, as demonstrated by the contamination case study presented within this article, the benefits of the technology far outweigh the effort required for the validation process. Increased industry communication on validation approaches, combined with the impetus of the PAT initiative, should enable more of these rapid microbiological methods to be successfully validated for use in the future. Finally, as rapid microbiology vendors and customers develop a deeper understanding of the nuances of each rapid system and communicate these insights with regulatory personnel, all microbiology laboratories undertaking these projects will accomplish more streamlined and successful validation and implementation.
Acknowledgements
The ScanRDI™ validation and implementation initiative at Wyeth Biotech has involved a cross functional team including personnel from Quality Assurance, Quality Control, Regulatory Affairs, Manufacturing, Product and Process Development, and BioPharmaceutical Analysis Microbiology. Specific thanks for support and input on the project go to Mary Griffin and Peri Ozker in Quality Control and Timothy Charlebois, James McColgan and Jason Gale in Product and Process Development. All experiments presented in this article were performed by Amelia Dominique and Kevin Luongo in the Quality Control Microbial Science and Technology Laboratory.
Information in this article is excerpted from ‘The Encyclopedia of Rapid Microbiological Methods’ published by PDA and DHI. All rights reserved.


References
- Chapter <1225>, Validation of Compendial Methods, U.S. Pharmacopeia, USP 27, United States Pharmacopoeial Convention, Inc. Rockville, MD. 2005.
- ICH Q2A. International Conference on Harmonization; Guideline on Validation of Analytical Procedures: Definitions and Terminology. Federal Register, 1995. Vol 60:40.
- ICH Q2B. International Conference on Harmonization; Guideline on Validation of Analytical Procedures: Methodology. Federal Register, 1995. Vol 62:96.
- Technical Report No. 33, Evaluation, Validation and Implementation of New Microbiological Testing Methods. PDA Journal of Pharmaceutical Science and Technology, supplement TR33, vol. 54, no. 3 (2000)







