Biomarkers of efficacy and safety
Posted: 22 August 2005 | | No comments yet
This article reviews the types of biomarkers currently available and approaches to discovering new biomarkers.
This article reviews the types of biomarkers currently available and approaches to discovering new biomarkers.
This article reviews the types of biomarkers currently available and approaches to discovering new biomarkers.
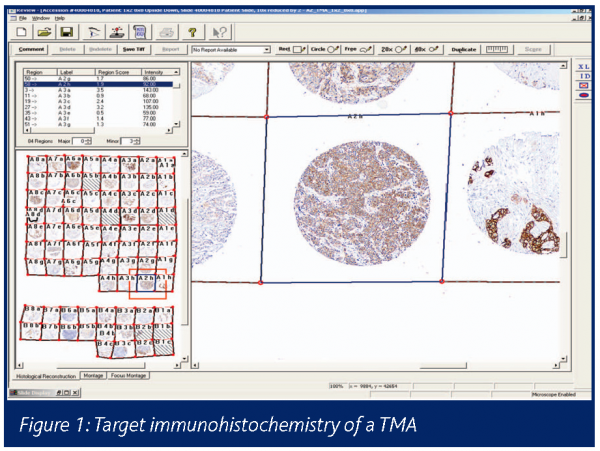
The category of type 0 biomarkers refers to pre-existing factors that influence disease susceptibility, outcome or therapeutic response (efficacy or toxicity) (Downing 2000). These markers may be genetic or acquired during life (e.g. viral infection, mutations in cancer) and constitute risk factors for disease. They equally form the basis of animal disease models, e.g. Zucker diabetic rats. The completion of the human genome and ongoing identification of the genetic polymorphisms and mutations within the germ line and cancer tissues has confirmed the previously unidentified existence of many patient subsets (Bertucci 2003). Consequently target selection for drug discovery has become more complex and the diagnostic challenges of personalised medicine and acquisition of human tissue banks from ethical and consented sources, compliant with the UK Human Tissue Act 2004, are now a priority for drug discovery. Personalised medicine is also a challenge for the commercial viability of niche new medical entities (NMEs) (Ross 2003, Lesko 2004). If clinical trial recruitment can be selectively based on patient subsets expressing the drug target, a better efficacy response could be anticipated, increasing the power of the clinical trial and attaining evidence for efficacy more rapidly as less patients need to be recruited. However smaller trials may impair the safety profiling of an NME. The technical means of screening hundreds of patient disease material samples (biofluids or tissue biopsies) is highly dependant on reagents (in particular antibodies) and tissue samples with reliable quality and provenance. The creation of tissue microarrays (TMAs) of clinical diagnostic biopsies and immunohistochemical staining for target protein expression allows for both the overall (brown) intensity of expression and heterogeneity of expression (e.g. cancer cell versus stroma, see Figure 1) and can enable early selection of target patient populations and individual cases.
However, as shown by the gefitinib experience in non-small cell lung cancer, additional complications of mutated targets influencing outcome will also modify the power of the trial (Simon 2004). Although having attracted less attention outside the arena of drug metabolism and pharmacokinetics (DMPK), patient genotype and target expression on bystander normal tissues is likely to help explain the heterogeneity in adverse events (AEs) in patients.
Type 1 efficacy biomarkers
This category refers to early events in disease progression, toxic changes or pharmacological events. As such it is of particular interest in preclinical disease models, toxicology and clinical pharmacology. The advent of the FDA Critical Path exploratory IND (http://www.fda.gov/oc/initiatives/ criticalpath/whitepaper.html) and Fast Track (http://www.fda.gov/cber/gdlns/fsttrk.pdf) and the EMEA New Safe Medicines Faster programmes has revitalised the search for new early and specific type 1 biomarkers as a means to expedite early clinical drug development. To deliver early readouts of pharmacological activity, clinical pharmacologists base decision points as follows:
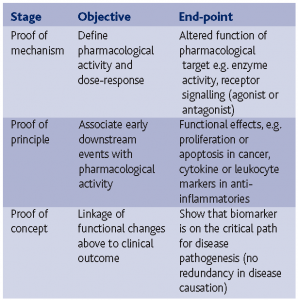

The other key requirement is to establish a pharmacokinetic:pharmacodynamic (PK:PD, biomarker based) relationship to assist with dose selection in normal volunteers and patients.
Generation of comprehensive adsorption, distribution, metabolism and excretion profiles in man has traditionally required the use of radiolabelled compounds and the regulatory safety package for such compounds. The advent of accelerator mass spectrometry (AMS) has permitted the early administration of trace (sub-pharmacological) doses of NMEs to man at microdoses of radioactivity not requiring lengthy radiobiological safety studies. Recent results from the CREAM programme have confirmed in a majority of cases that microdosing kinetic data are meaningful for PK at therapeutic exposures.
Type 1 safety biomarkers
Clinical adverse events and mortality related to drug treatment continues to be a major concern and the FDA has indicated that 1,500 plus subjects are needed to adequately define a drug’s safety profile prior to NDA submission. During clinical drug development, around 40% of the drug development attrition of NMEs is related to safety (20% clinical AEs, 20% preclinical toxicology) compared to 22% failing because of lack of efficacy. Biomarkers for toxicity in animals and man therefore merit greater attention.
Toxicity biomarkers should be;
- Early so that onset during dose escalation in early clinical trials can allow dosing to be reduced or stopped before clinically significant AEs occur.
- Specific so that the target organ indicated is accurate and erroneous signals from unrelated organs do not interfere with toxicity identification.
- Sensitive so that low doses causing toxicity can be detected so that high dose potentially irreversible injury is not caused.
Unfortunately, most clinical biomarker work has focussed on efficacy and drug metabolism and pharmacokinetic factors and there are many clinical target organ toxicities that either cannot be monitored, or fail to meet the three criteria above. Of particular concern are toxicities affecting target organs where component cells are terminally differentiated in adulthood, e.g. neurons, cardiomyocytes. Human patient AE rates may be insufficient for detection prior to marketing approval and withdrawal of marketed products can have a huge impact on patients, clinicians and pharmaceutical companies, e.g. coxibs. Human genetics have been associated with susceptibility to xenobiotic toxicity and carcinogenicity (Norppa, 2003) for many years and now with the advent of cytochrome P450 CYP polymorphism testing reagents, it should be feasible to exclude poor or atypical metabolisers from specific drug treatments.
Toxicity biomarkers used for standard safety screening in toxicology and during clinical development have changed little in recent decades. For example ‘liver function tests’ such as ALAT measure leakage of enzyme from hepatocytes rather than provide actual data on organ function. Safety biomarkers may be derived from hypothesis driven (‘biased’) approaches if a mechanism basis is known (Expert Working Group, 2005) but if mechanisms are unknown, more holistic ‘omics (‘fishing’) approaches may be needed to generate mechanism and biomarker data. More recent application of metabonomics (Nicholson 2002, Gavaghan, 2000) and transcriptomics can generation vast datasets requiring complex pattern recognition algorithms to provide a toxicity ‘fingerprint’. No single analyte to put into high throughput assays may emerge as a specific biomarker and altered multifactorial patterns may be needed. The problems of data reduction from meta-analysis of combined ‘omics technologies emerging from systems biology approaches becomes even more abstract although biological plausibility can be increased by pathway mapping to rationalise datasets (Wetmore 2004).
Type 2 biomarkers and surrogate biomarkers
This category refers to later clinical responses, either therapeutic or adverse events and offers the potential for early NDA approval in the basis of a validated surrogate biomarker approval in serious or life threatening disease. These biomarkers can become surrogates for traditional clinical outcomes based on morbidity or mortality. Where the time to end-point is protracted, e.g. cancer survival, surrogate biomarkers become attractive but demonstration that the surrogate is inextricably linked to survival can be challenging (Schatzkin 2001). This can however allow early Critical Path registration in serious or life threatening disease indications.
Approaches to discovery of new biomarkers
Biomarkers have been discovered during the course of many years as part of the laboratory investigation of clinical disease. Historically, clinical pathologists have found diagnostic biomarkers through a process of serendipity and hypothesis testing. One issue with the adaptation of these traditional biomarkers of disease is that they have been developed for clinical diagnosis where disease is already established and specificity has been paramount. Early onset and high sensitivity have been secondary considerations. They have been designed for target organ specificity rather than mechanism based. Also mechanism-based biomarkers can be disadvantageous in safety screening as opposed to hypothesis testing studies. Independence of mechanism is an advantage in many regards but will be less indicative of early adverse pharmacological or physiological consequences of drug treatment.
Proteomics traditionally has relied on the low throughput 2-D gel electrophoresis technology and can resolve not only individual proteins but also their post-translationally modified forms (Kennedy 2002, Man 2002, Srinivas 2002) and picking out single spots for tryptic digestion and sequencing by MALDI profiles permits both isolation and identification of novel biomarkers.
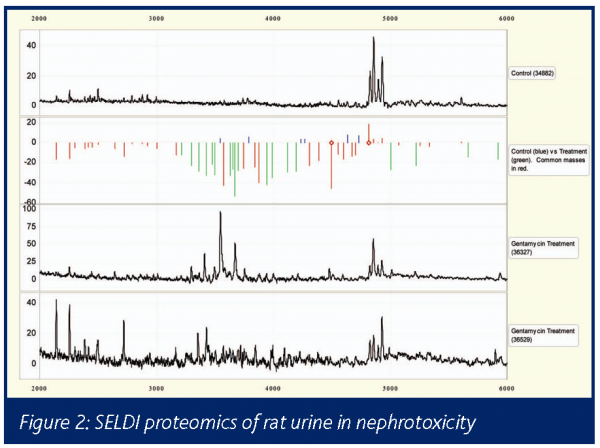
SELDI (Reddy 2003) provides a high throughput low sample requirement screening tool based on mass spectrometry proteomics (Petricoin 2002). Applications in preclinical toxicology are also numerous. For example, in a rat gentamycin nephrotoxicity model comparing control (top trace) and two treated (lower 2 traces) urine samples showed a spectrum of protein changes with each molecular weight altered expression being indicated by the green bars (Figure 2).
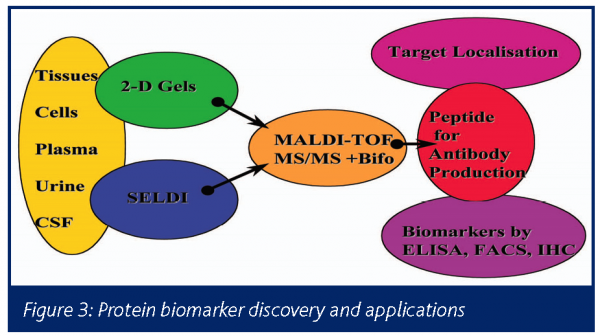
Once protein peaks or spots of interest have been identified as being of interest as a potential biomarker, it will be necessary to establish a more high throughput assay so the many experimental variables can be quantified. This is usually dependant on the development of an ELISA or similar assay. For example the protein or peptide of interest needs to be purified and sequences and antibodies generated if not commercially available, using the following schema (Figure 3):
Functional biomarkers e.g. PET (Frank 2003) reflecting target organ metabolic capability as opposed to downstream consequence such as cell leakage, death, proliferation or neoplasia provide a greater challenge than measurement circulating or excreted leakage markers in biofluids or cells. Imaging technologies have improved significantly in terms of speed, cost and resolution.
Structural biomarkers based in imaging can quantified, e.g. by volumetric measurement of articular cartilage, with great accuracy using MRI (Gandy 2005). Classical markers such as radiography have been developed into dual spectrum methods to quantify bone mineral density in trials of anti-osteoporosis agents
Novel biomarker validation ICCVAM process
The identification of novel biomarkers of efficacy or safety requires a lengthy process if they are to gain acceptance by regulations and the scientific and clinical community (Ransohoff 2004). In the US, the interagency ICCVAM organisation (http://iccvam.niehs.nih.gov/) recommends a number of stages for validation as follows:
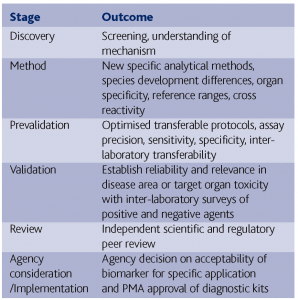

Biomarker deliverables
The following applications of type 0, 1 or 2 biomarkers to preclinical and clinical drug development should ensue;
a) Target localisation to specific cell populations involved in toxicity or disease pathogenesis
b) Target expression in potential toxicity target organs
c) Ensuring animal disease model is representative of pathogenesis in man
d) Refinement of pharmacodynamic endpoints
e) Biomarkers of efficacy can give earlier read-out of activity in animal models and identify human polymorphisms
f) Biomarkers of toxicity validated in animals can permit judicious dose escalation in man and early withdrawal if AEs occur
g) Biomarkers of toxicity can also help mechanism of action studies and species difference rationales expected by regulators and clinicians
Conclusions
Biomarkers are evolving rapidly and new ones are being discovered for both efficacy and safety applications. It is important that these are selected and validated in the appropriate settings and that information sharing permits regulatory and scientific acceptance of novel markers.



References
Bertucci, F, Viens, P et al (2003) DNA arrays in clinical oncology; promises and challenges. Lab Invest 83;305-316
Downing, G L (Ed) (2000) Biomarkers and surrogate endpoints; clinical research and applications. Elsevier. ISBN 0-444-50316-1
Expert working group on drug-induced vascular injury (2005) Drug induced vascular injury – a quest for biomarkers. Toxicol Appl Pharm 203;62-87.
Frank, R and Hargreaves, R (2003) Clinical biomarkers in drug discovery and development. Nat Rev Drug Disc 2;566-580
Gandy, S J, Brett, A D, Dieppe, P A, Keen, MC, Maciewicz, R A, Taylor, C J, Waterton, J C and Watt, I.(2005) Measurement of cartilage volumes in rheumatoid arthritis using MRI. Br J Radiol 78; 39-45.
Gavaghan, CL, Holmes, E.et al (2000) An NMR-based metabonomic approach to investigate the biochemical consequences of genetic strain differences; application to the C57BL10J and Alpk:ApfCD mouse. FEBS Letters 24251;1-6
Kennedy, S (2002) The role of proteomics in toxicology: identification of biomarkers of toxicity by protein expression analysis. Biomarkers 7;269-290.
Lesko, L J and Woodcock, J (2004) Translation of pharmacogenomics and pharmacogenetics; a regulatory perspective. Nat Rev Drug Discov 3;763-769.
Man, W J, Whire, I R et al (2002) Protein expression analysis of drug mediated hepatotoxicity in the Sprague-Dawley rat. Proteomics 2;1577-1585.
Nicholson, J.K., Connelly, J. et al (Feb 2002) Metabonomics: a platform for studying drug toxicity and gene function. Nature Reviews: Drug Discovery 1;153-161.
Norppa,H (2003) genetic susceptibility, biomarker responses and cancer. Mutat Res 544;339-348.
Petricoin,E F , Zoon,Z C et al (Sept 2002) Clinical proteomics: translating benchside promise into bedside reality. Natures Rev Drug Discovery 1;683-695.
Ransohoff, D F (2004) Rules of evidence for cancer molecular-marker discovery and validation. Nat Rev Cancer 4:309-314.
Reddy, G and Dalmasso E A (2003) SELDI ProteinChip array technology: protein based predictive medicine and drug discovery applications. J Biomed Biotech 4:237-241.
Ross, J S and Ginsberg, G S. (2003) The integration of molecular diagnostics with therapeutics. Amer J Clin Pathol 11;26.
Schatzkin, A and Gail, M (2001) The promise and perils of surrogate endpoints in cancer research. Nat Rev Cancer 2:1-9.
Simon, R and Maitournam, A (2004) Evaluating the efficiency of targeted designs for randomised clinical trials. Clin Cancer Res 10; 6759-6763
Srinivas, P.R., Verma, M.et al (2002) Proteomics in cancer biomarker discovery. Clin Chemistry 48;1160-1169.
Wetmore, B A and Merrick, B A (2004) Toxicoproteomics: proteomics applied to toxicology and pathology. Toxicol Pathol 32;619-642.




