The current challenge
Posted: 22 August 2005 | | No comments yet
We are currently living in an exciting age, where for the fist time ever, human diseases are being understood at a molecular level. Protein crystallography plays a major role in this understanding because proteins, being the major machinery of living things, are often the targets for drugs.
We are currently living in an exciting age, where for the fist time ever, human diseases are being understood at a molecular level. Protein crystallography plays a major role in this understanding because proteins, being the major machinery of living things, are often the targets for drugs.
We are currently living in an exciting age, where for the fist time ever, human diseases are being understood at a molecular level. Protein crystallography plays a major role in this understanding because proteins, being the major machinery of living things, are often the targets for drugs.
The function of these proteins is determined by their three-dimensional structures hence a detailed understanding of protein structure is essential for rational design of therapeutic treatments (Blundell et al 2002).
The most powerful method for determining the structure of proteins is X-ray crystallography which is totally reliant on the availability of high quality crystals.
Protein crystallisation is a complex multi-parametric process. In addition, most proteins of interest are limited in supply and are labour-intensive and costly to produce. Structural proteomics, which aims to determine the structures of thousands of proteins, has put great pressure on the crystallography community to produce suitable crystals. As a result, crystallisation is gathering a new momentum as evidenced by the increased interest and investment of pharmaceutical companies in crystallisation equipment and expertise, a high demand for practical courses in crystal growth and the increasing numbers of commercial companies selling crystallisation kits and tools.
Current state-of-the-art
The past four years have seen some of the greatest achievements in the field of protein crystallisation. It is now feasible to screen thousands of potential crystallisation conditions by dispensing trials consisting of nanolitre volumes in a high throughput mode. This has cut the time of setting up experiments from weeks to minutes, a scenario that was unimaginable a few years ago. Even more incredible, is the revelation that diffracting crystals can be produced from protein samples in volumes as small as 5-20 nanolitre (Hansen et al, 2002; Stevens, 2000). The subsequent phase of image capture and analysis of the crystallisation drops is also progressing in great strides (Luft et al 2003; Klei, 2004, Bard, 2005).
However, in spite of the impressive advances accomplished, the crystallisation problem has not been solved. High throughput has not yet resulted in high output.
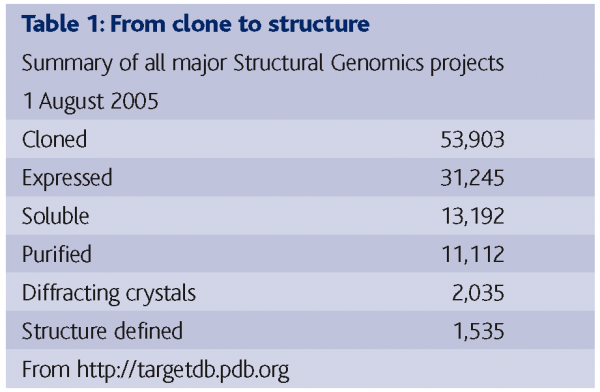
Table 1 shows the sum of results from 17 structural genomics/proteomics projects worldwide on 1 August 2005, demonstrating the success rate of getting from cloned protein to structure determination (The numbers for each individual project vary as they are dealing with different genomes and have started at different times). The results demonstrate that out of 53,903 clones and 31,245 expressed proteins, 13,192 proteins were soluble and 11,112 were purified. Of these, 2,035 good crystals were produced leading to the determination of 1,535 structures. It transpires that more than half of the trials that were set up from pure soluble proteins had produced crystals, but only about half of those crystals were diffracting ones. It was initially expected that if proteins could be made soluble and highly pure, there would be no problem in getting them to crystallise. However, this is not the case. The results are indicating that even when proteins can be expressed and rendered highly pure and soluble, this still does not guarantee a yield of useful crystals (Chayen, 2003). Some proteins (referred to as ‘low hanging fruit’) do crystallise during the initial screening stage but many trials are yielding micro crystals or low-ordered ones.
The conversion of crystals into useful ones requires the design of optimisation techniques that go beyond the usual fine-tuning of the initial conditions. In the rush towards structural genomics/proteomics, such optimisation techniques have been somewhat neglected, mainly because it was hoped that large-scale screening would be sufficient to produce the desired results. In addition, optimisation had relied on what is known as the ‘black art’ of crystallisation and in particular instances, more complicated methods that are often difficult to automate and to adapt to high throughput. There is now an urgent need to find ways to simplify, automate and miniaturise these techniques in order to cope with the vast numbers of ‘leads’ resulting from the screening procedures.
The next sections highlight several optimisation methods that have been miniaturised and automated in our laboratory so that they can easily be applied by any lab conducting automated crystallisation trials. These methods are applied in cases where standard optimisation by fine-tuning of the screening conditions has failed to produce high quality crystals. The methods involve active influence and control of the crystallisation environment while the trial takes place, in order to lead crystal growth in the direction that will give the best results.
Crystallisation strategies and techniques
Crystallisation is a phase transition phenomenon. Crystals grow from an aqueous protein solution when the solution is brought into supersaturation (Ataka, 1993). Supersaturation is achieved by varying the concentrations of precipitant, protein and additives, pH, temperature and other parameters (McPherson, 1999; Bergfors, 1999; Ducruix and Giege, 1999).
Crystallisation proceeds in two phases, nucleation and growth. Nucleation, which is a pre-requisite and the first step in crystallisation, requires different conditions than those of growth. In an ideal experiment, only a few nuclei form, then turning into well-ordered crystals. However, most crystallisation trials are not ideal and more often than not, excess nucleation occurs, whereby numerous clusters of tiny useless crystals are formed. The task of the experimenter is to optimise the crystals by controlling the nucleation step and/or by separating the phases of nucleation and growth.
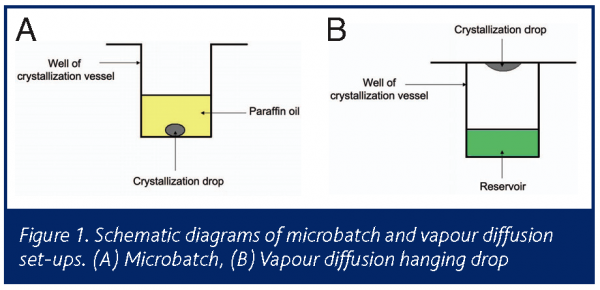
The two most common methods of crystallisation are vapour diffusion and microbatch (Figure 1). The free interface diffusion is recently gaining more popularity than it has had in previous years. All three methods are fully automated and operate in high throughput mode for screening and fine- tuning of conditions.
Microbatch is a micro/nano scale batch crystallisation that involves mixing of protein and the crystallising agents at conditions that aim to achieve supersaturation immediately upon mixing. The trials are dispensed and incubated under paraffin oil in order to prevent the evaporation of such small volumes (Figure 2a). This is in contrast vapour diffusion (and all other crystallisation methods based on diffusion) that are dynamic systems in which the protein solution is undersaturated at the outset of the experiment and gradually reaches supersaturation by equilibration with a reservoir solution which contains the crystallising agents (figure 2b). Microbatch is therefore especially useful for optimisation by controlling the crystallisation environment. This is because in batch, conditions are known and constant, making it easier to follow the history of the sample and achieve reproducibility.
The microbatch method (Chayen et al 1990) involves the application of a layer of paraffin oil, thick enough to render evaporation through it negligible within the time scale of a crystallisation experiment (typically one week to one month). The thickness is 4mm, which translates to 6ml in standard microbatch plates, or a ratio of 1 to 50 between the drop and the oil. A thinner layer of paraffin oil will allow evaporation leading to drying of the drops. Other oils such as silicone oil or mixtures of paraffin and silicone oils also allow evaporation of the trials (D’Arcy et al, 2004). The different properties of the oils facilitate the control of crystallisation.
Automated optimisation techniques
Decoupling of nucleation and growth
The most effective moment to intervene with a crystallisation experiment is soon after the formation of the first critical size nuclei which will eventually form the crystal.
A simple means of keeping the nucleation at an optimal level is by starting the trials at nucleation conditions and after a given time ‘backing off’ to conditions of growth. The conditions for nucleation (i.e. conditions that would produce the low quality crystals if the trials were left undisturbed) are found by means of an automated system and drops are set up as microbatch trials under these conditions. At various time intervals after set-up of the experiments, the robot is programmed to automatically insert the dispensing tip into the drops and add buffer or protein solution, thereby diluting the trials. Single diffracting crystals were routinely attained, equivalent to the best, very rarely obtained without employing the dilution procedure (Saridakis et al., 1994; Saridakis et al 2002; Chayen, 2005).
Control of evaporation kinetics
Dilution experiments involve revisiting the crystallisation drops which may cause disruption to the trial. An alternative way of limiting the nucleation without touching the trial drop can be achieved by inducing nucleation and then stopping it before it becomes excessive. This is achieved by controlled evaporation and therefore concentration, of the drops through a thin oil layer. Evaporation is later arrested by increasing the thickness of the oil layer (Chayen and Saridakis, 2002). Experiments are set up using a robot which dispenses the trials under the lower volume of oil. The robot is then programmed to add oil at various time intervals after setting up the trials. If trials were allowed to evaporate without arresting, the drops would dry out. By arresting evaporation in the early stages of nucleation, the result is the formation of fewer crystals of better quality.
Determination of the time of dilution or arrest of the trials
Dilution or the arrest of nucleation by addition of oil must be performed before crystals are visible. The time is therefore selected by reference to the time in which it took to see the first crystals in the initial screens. For example, if crystals appeared within 24 hours, nucleation would have occurred anytime between setting-up the experiments to several hours before the crystals appear. Hence the dilution/arrest should be done at intervals of 1-2 hours after set up. If crystals appear after 4 days, dilution should be performed at intervals of 8-12 hours. Trials that are diluted/arrested too soon will produce clear drops while those that are too late will yield low quality crystals.
The exact time for intervention can be pinpointed precisely by following the trials using dynamic light scattering (DLS). DLS is sensitive to variations in particle size in the range of approx. >1nm and interactions of protein molecules in solution (e.g. Schmitz, 1990; Mikol et al 1990; Malkin et al 1993; Schueler et al., 1999). It is therefore a useful tool for an early, non-invasive, in-situ observation of a crystallisation event. Experiments have reported that the time at which DLS showed a significant change in the size-distribution profile of species in solution, corresponded to the time at which the solution was effectively diluted to growth conditions, for optimal growth (Saridakis et al 2002). To date DLS experiments are performed semi-automatically but several companies are currently working on fully automating, as well as miniaturising the process using the microbatch and vapour diffusion methods of crystallisation.
Dilution can be achieved using all crystallisation methods. In the case of vapour diffusion, either the drops themselves or the reservoirs can be diluted. However, microbatch is still the simplest and most amenable to automation of such a procedure.
Crystallisation in gelled media
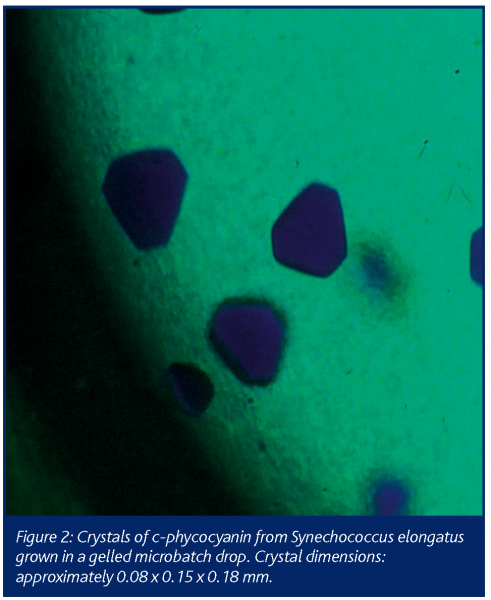
The quality of crystals can be significantly improved when grown in gel media. Growth in gel which reduces both convection of the protein molecules, thus favouring their slow diffusion to the crystal faces and sedimentation of the growing crystals, in some way mimicking a microgravity environment (Robert et al 1999 and references therein). Crystallisation in gels also presents an additional means to promote or inhibit nucleation depending on the type of gel applied (Cudney et al 1994; Sica et al 2002). Although crystallisation in gels can be performed in all methods of crystallisation, this technique remains underused. This may be due to the relatively complicated procedures required when applying gels to crystallisation trials and also to the large quantities (mostly over 10 ml) of sample needed. The last three years have seen major improvements in the use of gels by way of miniaturisation and automation. The Granada Crystallization Box (Garcia-Ruiz et al., 2002) enables the growth of crystals in 1μl volumes of gel inside capillaries. An even simpler way, is to set up gelled trials in microbatch, which for the first time, has enabled the automatic dispensing of sub-microlitre gelled drops in a high throughput mode (Chayen and Saridakis, 2002). The experiments are performed using a computer-controlled dispensing system in which precipitant, buffer, protein and additives are put into different syringes. By placing a gel solution (preferably tetramethyl orthosilane (TMOS)) while it is still in liquid form, into one of the syringes, it is possible to automatically dispense microbatch trials which form the gel/crystallization mixtures in final volumes of 0.3 – 2 μl. This can be achieved during a comparable time-scale and with the same ease as conventional automated microbatch trials. Figure 2 shows a picture taken under the microscope of crystals of c-phycocyanin from Synechococcus elongatus grown in a gelled microbatch drop under paraffin oil. Comparison of crystals grown in gelled drops with those grown in standard trials demonstrates a significant improvement in crystal order.
Future prospects
Obtaining high quality crystals is becoming increasingly crucial to progress in the post genomic era. Whether in the academic laboratory, the pharmaceutical industry or as part of structural genomics projects, it is always vital to have a portfolio of crystallisation techniques that can be applied, especially in the cases of proteins that are proving difficult to crystallise. In order to be useful to the structural genomics efforts, it is also important to miniaturise and automate as many techniques as possible with a view of adapting them to high throughput mode.
Control of crystallisation kinetics, dynamic separation of nucleation and growth and crystallisation in gels can now be performed automatically while using nanolitre or microlitre volumes of sample. These methods provide a variety of avenues to be explored either in parallel to conventional methods, or when screening and subsequent fine-tuning of the initial screening conditions have failed to produce high quality crystals.
The post genomics era has opened up the scope for the development of new techniques and tools to overcome the bottleneck of protein crystallisation. Sophisticated instrumentation is currently being designed and implemented for the screening process of crystallisation. Optimisation techniques have not yet caught up with these advances but effort is now invested into this area. Thus the coming years promise to bring advances in the more complicated optimisation techniques that will play a major role in raising the success rate of producing high quality crystals, and will equip the genome project to deal with its awesome task.



References
Ataka M. (1993) Protein crystal growth: an approach based on phase diagram determination. Phase Transitions 45, 205-219.
Bard, J. (2005) European Pharmaceutical Rev. 1, 40-45.
Bergfors, T.M. ed 1999. Protein Crystallization: Techniques, Strategies, and Tips.
International University Line, La Jolla.
Blundell TL, Jhotti H, Abell C. (2002) High-throughput crystallography for lead discovery in drug design. Nature Rev Drug Discovery 1, 45-54.
Chayen N.E. (2003) Protein Crystallisation for Genomics: Throughput versus Output J. Structural and Functional Genomics 4, 115-120.
Chayen, N., Shaw Stewart, P.D., Maeder, D.L. and Blow, D.M. (1990). An automated system for microbatch protein crystallization and screening. J. Appl. Cryst. 23, 297-302.
Chayen, N.E. (2004) Turning protein crystallization from an art into a science. Curr. Op. Struct Biol. 14, 577-583.
Chayen, N.E. (2005) Methods for separating nucleation and growth in protein crystallization Progress in Biophysics and Molecular Biology 88, 329-337.
Chayen, N.E. and Saridakis, E., 2002, Protein Crystallization for Genomics: Towards High-Throughput Optimisation Techniques. Acta Cryst. D 58, 921-927.
Cudney, B., Patel S, and McPherson, A. (1994). Crystallization of macromolecules in silica gels. Acta Cryst. D 50, 479-483.
D’Arcy, A., Mac Sweeny, A. and Haber, A. (2004) Practical aspects of using the microbatch method in screening conditions for protein crystallisation. Methods 34, 323-328.
Ducruix A, Giegé R ed: Crystallization of Nucleic Acids and Proteins, A Practical Approach, edn 2. Oxford: Oxford University Press; 1999.
García-Ruíz JM, Gonzalez-Ramirez LA, Gavira JA, Otálora F (2002) Granada Crystallization Box: a new device for protein crystallization by counter-diffusion techniques. Acta Cryst D 58, 1638-1642.
Hansen, C. L. Skordalakes, E., Berger, J. M., & Quak, S. R. (2002) A robust and scalable microfluidic metering method that allows protein crystal growth by free interface diffusion PNAS 99, 16531-16536.
Klei, H.E. (2004) European Pharmaceutical Rev. 3, 85-88.
Luft, JR, Collins RJ, Fehrman NA, Lauricella, AM, Veatch CK, DeTitta GT (2003). A deliberate approach to screening for initial crystallization conditions of biological macromolecules. J Struct Biol 232:170-179.
Malkin, A.J., Cheung, J. and McPherson, A. (1993) Crystallization of satellite tobacco mosaic virus 1. nucleation phenomena. J. Cryst. Growth 126, 544-554.
McPherson, A. 1999. Crystallization of Biological Macromolecules. Cold Spring Harbor Laboratory Press, Cold Spring Harbor.
Mikol V, Hirsch E, Giege R. (1990). J. Mol. Biol. 213, 187 – 195.
Robert, M.-C., Vidal, O., Garcia-Ruiz, J.-M. & Otálora, F. (1999) In Crystallization of Nucleic Acids and Proteins. (Ducruix, A. & Giegé, R., eds.), pp.149-175, Oxford University Press, Oxford.
Saridakis, E., Dierks, K., Moreno, A., Dieckmann, M. W. M. and Chayen, N.E. 2002, Separating Nucleation and Growth in Protein Crystallization Using Dynamic Light Scattering. Acta Cryst D 58, 1597-1600.
Saridakis, E.E.G., Shaw Stuart, P.D., Lloyd, L.F. and Blow, D.M., 1994. Phase diagram and dilution experiments in the crystallization of carboxypeptidase G2. Acta Cryst. D 50, 293-297.
Scheuler J, Frank J, Saenger W, Georgalis Y, 1999. Thermally induced aggregation of human transferring receptor studied by light-scattering techniques. Biophysical Journal 77, 1117 – 1125.
Schmitz S K, 1990. An introduction to dynamic light scattering by macromolecules. New York: Academic Press.
Sica, F., Demasi, D., Mazzarella, L., Zagari, A. Capasso, S. Pearl, L.H., D’Auria, S., Raia, C.A. and Rossi, M. (1994). Elimination of twinning in crystals of Sulfolobus solfataricus alcohol dehydrogenase holo-enzyme by growth in agarose gels. Acta Cryst. D 50, 508-511.
Stevens, R. (2000) High throughput protein crystallization Curr. Opn. Struct. Biol.10, 558-563.




