Versatile miniaturised HTS
Posted: 7 March 2005 |
Research and development for a pharmaceutical company is a difficult and lengthy process. It stretches from the discovery phase to preclinical and clinical development stage, through the drug approval period ultimately to clinical application. The discovery research phase is one of the early key processes. The research starts with target identification and validation. Assays are […]
Research and development for a pharmaceutical company is a difficult and lengthy process. It stretches from the discovery phase to preclinical and clinical development stage, through the drug approval period ultimately to clinical application. The discovery research phase is one of the early key processes.
The research starts with target identification and validation. Assays are then developed to measure biological activity and to run a high throughput screening (HTS) study to generate a first hitlist of active compounds. Identification of active compounds results in lead structures that are then optimised to drug development candidates. Pharmaceutical R&D can be viewed as a continuous struggle against the high attrition rate of active compounds in the pipeline. Growing numbers of compound libraries to be tested in a rapidly increasing number of new and complex bioassays calls for experimental tools optimised in respect to sensitivity, speed and statistical accuracy. HTS is regarded as a powerful tool to keep pace with these demands in pharmaceutical research. However a major challenge is to maintain a high quality per data point, even under miniaturised assay conditions and in a high level of automation and fast turnover of different projects.
Technology
Compound transfer HTS at Novartis is organised into centres of excellence dealing with dedicated HTS tasks based on technology or target platforms. The Novartis NanoScreening facility is running HTS studies in highly miniaturised high density plate formats (1536w or 2080w) on two different screening modules (Evoscreen Mark-II and Mark-III) with two different compound reformatting strategies on three independent modules. Total assay volumes range from 0.8 to 6 µl per well or assay data point. This level of assay miniaturisation requires compound handling in the low nanoliter range. Two different strategies are applied in the Novartis Nanoscreening facility. The Evoscreen MITONA (micro to nano, Evotec Technologies)1,2 with its piezo-driven pipettes is dedicated to compound reformatting from 384w source plates into 2080w format in the working range of 10 to 200 nl. The working range of compound reformatting from 384w compound source plates into 2080w assay plates on the MITONA is 10 to 100 nl. Most of the assays in the 2080w format are run with the Nanostore strategy. With the Nanostore concept the compounds are reformatted into the assay plates and dried. Typically those plates are prepared in advance and stored in the 2080w plates (Nanostore). Unattended operation of the MITONA can achieve the reformatting of >100.000 compounds per 24 h. The final HTS is then run in the absence of DMSO and additives assist complete dissolution of the compounds in the final assay buffer. For 1536w screening another strategy is used. In collaboration with Zinsser Analytics, two automated compound reformatting modules were developed for rapid compound transfer from 384w source plates into 1536w assay plates with the Hummingbird capillary effect technology (Genomic Solutions). Due to the high speed of operation, the assay plates containing the compounds can be generated on demand and ad hoc. The working range of compound reformatting from 384w compound source plates is 25, 50, and 100 nl. The unattended operation of one module can achieve the reformatting of >150.000 compounds per 13 h. This strategy allows screening of freshly reformatted compounds. Both strategies are well developed to enable reliable compounds transfer in the low nanoliter scale with very low outlier rates (< 0.1 %) and CVs (coefficient of variation < 8%). The development of efficient pipette washing protocols brought the well-to-well cross contamination rate below 0.1%.
Assay screening
Full flexibility in handling a great variety of HTS targets is also maintained with the setup of the two screening modules. The Evoscreen Mark-II and Mark-III SCARINAs (Screening and Reading in Nanoscale, Evotec Technologies) can be dedicated to different and mostly independent HTS campaigns. The Mark-II SCARINA(1) is the confocal single molecule detection workhorse running assays in the 2080w format with total volumes of 0.8 to 1.6 µl per well or alternatively with 2.5 to 3.5 µl per well in the 1536w plate format. Assay steps can contain up to eight different reagent additions with piezo-driven NANOdispensers, including incubations in two independent high humidity incubators at independent temperature settings. All assay process steps are protected against evaporation of the very low assay volumes. Even total assay volumes down to 0.8 µl per well can be handled for up to 8 hours in the module without significant evaporation effects or patterns on nanotiter plates. The mainly unattended operation can achieve a throughput of >100.000 compounds screened in 24 h with a scheduling software organising the assay process steps (Bernstein, Evotec Technologies). The Mark-II is operating exclusively biochemical HTS studies with confocal assay readout applying single molecule detection technologies on two identical confocal readers. This generates high content assay readout with various assay parameters. Applied HTS readout technologies are FLINT (Fluorescence Intensity), 1D-FIDA (Fluorescence Intensity Distribution Analysis), 2D-FIDA (two-dimensional Fluorescence Intensity Distribution Analysis) and cFLA (confocal Fluorescence Lifetime analysis) and combinations thereof3. Advantages of confocal single molecule assay readout are discussed below. The second generation screening module, the Mark-III SCARINA2, has an even more open architecture than the Mark-II. The above mentioned piezo-driven nanoliter dispensing and confocal readout capabilities in 2080w and 1536w format of the Mark-II are complemented on the Mark-III with more classical assay readout technologies to broaden the field of assay applications. Standard 1536w plates (as defined by SBS, Society of BioMolecular Screening) can be used on the module as well as the 1536w or 2080w confocal readout plates. For efficient and fast reagent dispensing a SynQuad (Genomic Solutions) dispenser is implemented as well. On the basis of an open plug-and-play architecture, an Analyst GT multimode reader (Molecular Devices) is added to strengthen the assay portfolio towards more classical assay readouts, such as fluorescence intensity, TR-FRET (time-resolved fluorescence resonance energy transfer), luminescence, fluorescence polarisation or even absorbance. Those comparatively fast and robust readouts enables the throughput of >100.000 screened compounds per day or even more. The capability of 1536w suspension cell dispensing further opens the assay technology portfolio of the Mark-III towards cellular assays. The technology on both systems delivers high quality in miniaturised format. Even in nanoliter reagent dispensing over whole 1536w and 2080w plates in combined assay protocols, CVs of well below ten per cent are achieved. Altogether, both automated screening platforms – Mark-II and Mark-III – enable the Novartis Nanoscreening to run HTS studies in a miniaturised high density plate format with the capability to easily switch the assay format between different HTS studies. The decision of assay format choice is driven by scientific rationale during the assay planning or development phase, rather than based on the merely technological capabilities that are available. The portfolio of targets classes for applied biochemical HTS is broad. Main enzymatic target classes are kinases, phosphatases, proteases, as well as other enzymes. Ligand binding assays are performed for protein-protein interactions or for interactions between protein and ligands like DNA, RNA or low molecular weight ligands.
Confocal single molecule detection
Confocal single molecule detection (SMD) readout technique generates high content assay readouts with various assay parameters during one measurement. This offers flexibility and added value in assay data evaluation and enables investigation of a more extensive set of data during assay development. Complex binding events can be evaluated resolved on the different involved assay species (i.e. bound and unbound ligand), certain assay technologies become only feasible in the confocal setup (i.e. FIDA) 3, 4, 5, 6. Added value, like species, resolved molecular brightness or molecular concentration serve as quality indicators for the HTS results. The signal detected by a confocal single molecule detection reader setup fluctuates due to the diffusion of single fluorophore molecules into and out of the laser focus and detection volume, each causing a burst in fluorescence emission and, thus, in detected fluorescence photons. By analysing the resulting distribution of signal intensities, FIDA distinguishes the species of a sample according to their different values of specific molecular brightness and quantifies each species by the determination of its absolute concentration. 2D-FIDA is capable of measuring the rotational motion (fluorescence polarisation or anisotropy) of fluorescent assay systems resolved on molecular species signal. In addition to the FIDA performance, 2D-FIDA achieves additional molecular resolution by the concurrent determination of two specific brightness values originating from two detection channels. By observing the molecularly resolved fluorescence anisotropy, simple as well as more complex binding events and enzymatic reactions may be followed. Alternatively, such events may be resolved by labeling with two dyes that express different fluorescence emission bands. As a read-out, either two-colour excitation by different lasers or one-colour excitation using a FRET dye pair can be chosen3. Furthermore, FIDA and 2D-FIDA compensate for variable background signals, owing to scattering or autofluorescent impurities of compounds or their decomposition products, which is one of the major sources for screening artifacts. Confocal Fluorescence Lifetime analysis (cFLA) analyses the fluorescence decay of samples resolved in the very low nanosecond scale with a pulsed diode laser operating in the picosecond scale. This very fast and efficient kinetic readout is, in contrast to FIDA and 2D-FIDA, in a large detection range independent on fluorophore concentrations and can achieve very good assay performance.
HTS
Assay development and implementation Automated screening of sample libraries in the range of 1.000.000 compounds or even above per HTS project contains certain risks. High numbers of data points are generated in a very short period of time at relatively high costs and with little time to react and to optimise the setup if problems in assay performance arise (i.e. signal to background window too small or too much enzyme turnover). One of the most important phases of each HTS is its preparation and therefore the assay development and HTS implementation phase. To minimise the risks, certain experiments are key steps and go/no-go criteria for the actual automated screening campaigns. Biological and technical feasibility of the assay setup are established during the early assay development phase. In the majority of projects the decision of choosing a ‘classical’ or a ‘confocal’ assay readout technique is made in this phase. The decision is, in many cases, driven by the availability of assay components like protein or labeled ligands, which are often produced in complex eukaryotic expression systems with low yield. The unchallenged sensitivity of the confocal single molecule detection readout, especially in ligand binding assays, is balanced by the advantages of classical non- single molecule detection readout. Classical assay readout sometimes offers a broader range of assay signal above the single molecule detection conditions. This is often advantageous in enzymatic assay systems (i.e. high substrate concentrations or turnover). The downscaling and miniaturisation of the assay to 1536w or 2080w plate format and testing of reference compounds in the final assay setup are crucial. One of the key steps in the implementation phase is the final adjustment of assay parameters with a pilot screen. These assay parameters will have a great impact on assay results and on screening performance. Assay sensitivity for identification of weak modulators (i.e. weak inhibitors) is unfortunately often diminished by attempts to generate an assay readout that is ‘too stable’. Generally assay stability is often expressed by low deviations of the assay signal over whole assay plates or runs and high statistical parameters like Z and Z’-value7. However, it must be taken into consideration that the Z and Z’ values are statistical parameters with partly limited testimony of assay sensitivity towards the identification of active compounds. For example, typical steady-state enzyme assays are run under conditions that don’t always produce the best signal to background or Z’ values, in order to maximise the sensitivity to competitive inhibitors. Throughout the whole process a priority matrix for the applicable readout techniques helps to challenge and select the most feasible readout. Issues with values generated for reference inhibitors in respect to KM, Ki, KD or IC50 values for known actives or systems must be addressed. A key question is: “do we find weak inhibitors?” Stabilisation of the assay readout in miniaturised format with maximum sensitivity of the assay at good, but not perfect, Z and Z’ values are the crucial demands. On the other hand screening of 1.000.000 compounds or more has to be stable against assay artefacts (promiscuous inhibitors, false positives, autofluorescence, fluorescence quenchers, pipetting errors, reagent instability in process etc). To address these issues Novartis has implemented the strategy of testing a pilot or ‘pre-screen’ library before starting the full screening campaign. It can be divided into dedicated sub-parts for answering different questions. The majority of compounds are a statistical representation of the whole compound collection based on in silico diversity parameters (ca. 10.000 compounds). A number of known assay artifact compounds (ca. 100), based on several years of HTS readout experience, are added to challenge the assay against common screening issues like promiscuous inhibitors, false positives, compound autofluorescence, or compound fluorescence quenching effects. This pre-screen with a limited compound library helps to make hit rate assumptions for the actual HTS and to test stability and reproducibility of the screen. The figure below shows the influences of two different setups on some key assay parameters. The assay is tested with the pre-screen library applying two different ligands (setup A and B) in a protein-ligand binding 2D-FIDA assay in miniaturised assay format (2080w, 1.2 µl per well). Both assay setups have good performance in terms of Z’ values of 0.64 and 0.73 for setup A and B respectively. However, an overlay of the activity histograms for both setups shows that the activity distribution of setup A follows a much broader distribution than setup B. Separation of inactive from active compounds and a reliable selection of even weak active compounds is much easier with setup B than with setup A. A potential threshold for hit selection based on three times the standard deviation of the signal results in a threshold of 39.6% inhibition for setup A and 18.1% for setup B. This, of course, has an impact on the final hitlist that can be generated. Setup A will not reliably identify inhibitors with IC50 >10 µM, whereas setup B will be capable of finding even weak inhibitors with IC50 > 20 µM. Analysis of those compounds from the prescreen section, which are known to be artifacts in screening, showed a relatively high hit-rate with setup A (22 hits > 40% inhibition), whereas setup B showed a relatively low hitrate (4 hits > 40% inhibition) for those compounds. This clearly indicates that the number of false positives and also false negatives is much higher in setup A compared to setup B.
High throughput screening
After establishing robust and reliable HTS protocols and validation of the methods on automated screening modules, the actual HTS slot is tightly scheduled. In order to screen the whole compound deck under comparative and similar conditions, high numbers of compounds per day must be tested. Depending on the applied readout technology, a slot of 2-3 weeks of consecutive screening is dedicated to the primary HTS campaign. One of the key components of the automated HTS operation is automated data processing with online data monitoring to enable fast and efficient quality control. Automated alert functions and systems allow for unattended operation without an unacceptable high risk of occurring errors. The alert system is configured to handle a variety of system errors ranging from database connect or assay plate registration errors during reformatting until system errors on the screening module (i.e. plate handling errors, reagent reservoir level detection, system failures) and the automated assays readout quality control and nanotiter plate data acceptance or rejection by preconfigured assay definitions. For unattended operation, especially during night or weekend runs, the alert system is capable of transmitting an alert message via e-mail or SMS message. This setup enables a fast and efficient notification and error recovery by dedicated power users. Error recovery can happen via remote access, to control and modify the screening modules and is further supported with web camera visualisation of the laboratory. During the screen or afterwards, the user is operating with the screening data interactively. Key parameters of a screening campaign on plate-to-plate level, based on runs or even the whole HTS campaign, are visualised and the final assay readout and results are tracked per data point in the core database system. This visualisation and quality control system can allow for the visualisation of parameters as shown in the figure below. Examples are histograms of Z’ values and histograms of nanotiter plate throughput per run, as well as adaptive visualisation of a plate result plot showing the activity distribution over plates and libraries. Visualisation of the number of inhibitors or Z’ values per plate shown over time can serve as indicators of process problems and for later hitlist follow up and the selection of the primary hits.
Hit selection process
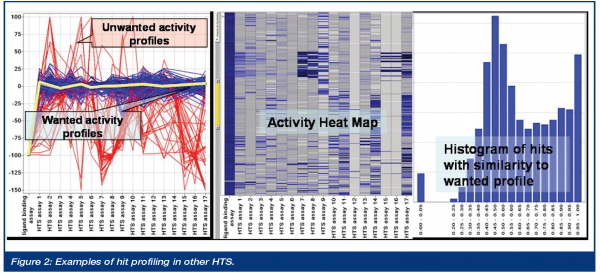
In order to select primary hits for IC50 validation follow up (dose response curves), historical HTS results are merged with corporate and logistical data about the screening samples. These data can be visualised for hit selection i.e. as a profile chart showing the activity of the primary hits in an adaptive set of other screens. The selection of the comparison screens is done individually per target to select i.e. selective inhibitors, or to rule out technology relevant frequent hitting samples. In other words, many seemingly active hits in the primary screen can be removed because they are promiscuous inhibitors, or do not have the correct selectivity profile against related targets. This process is especially important for HTS projects that must identify a low number of true positives from among a large population of false positives. The figure below exemplifies the procedure. The activity profile per sample in the actual HTS and in the set of other comparative screens should follow a certain pattern. Activity in the actual HTS and weak, or no, activity in the majority of the other screens is one example of the wanted profile of a sample. The yellow profile shows the optimum wanted activity profile. Blue profiles have high similarity to the wanted activity profile; red profiles have a strong deviation, which is most probably a sign for frequent hitter samples that should be excluded in this example. The level of activity in other screens is, of course, adaptive and a high number of screens counterbalances the risk of loosing attractive hits active in only one or few other screens. This process establishes generation of a primary hitlist of higher quality compared to a hitlist that is merely based on high activity in the given HTS target. The past has shown that the success rate of HTS hits can be increased significantly if this process is used, as even weak actives in primary screening can later evolve as good and attractive candidates for hit-to-lead activities. Further in-silico activity and selection is based on the hitrate and follow-up capabilities of the target.
Validation
After the selection process, the samples are retested in replicates in dose response curves to generate the IC50 values and a higher level of data quality than a single data point per sample as in the primary screen. Counter screens or secondary assays round up the overall picture of the validation hitlist and add information to facilitate the hit selection done in the hit-to-lead activities further downstream. Altogether the combination of modern automated screening technologies in miniaturised format merged with the capabilities of data handling and automated alert systems paves the road towards enhanced drug discovery processes in Pharma industry with high quality and fast and efficient project turnover.
Acknowledgement
We gratefully acknowledge the valuable input of all members from Novartis’ Discovery Technologies (DT) department at Novartis Pharma that either helped integrating the NanoScreening platform into the lead finding processes at Novartis during the past years or are still part of these processes, such as tool production, assay development, compound supply and data management. Furthermore, we greatly appreciate the continuous support and advice from numerous people at Evotec OAI and Evotec Technologies. We would like to thank Dr. Fraser Glickman and Dr. Lorenz Mayr for kindly reviewing the manuscript. 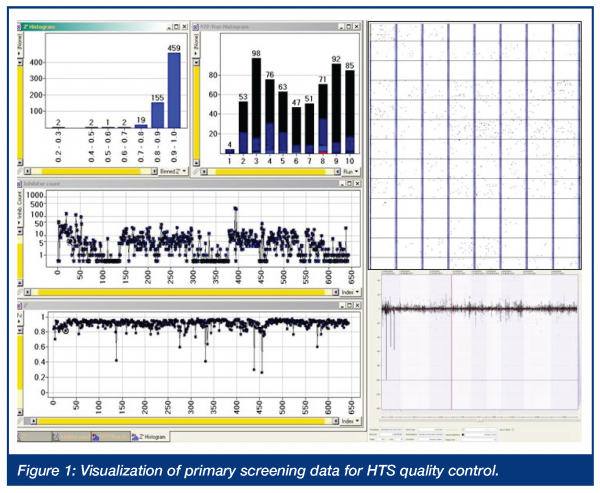


References
- Gentsch, J., Bruttger, O. High Throughput Screening at the Nano Scale. (2000), JALA, 5, 3, 60-65.
- Jäger, S., Garbow, N., Kirsch, A., Preckel, H.,Gandenberger, F.U., Herrenknecht, K., Rüdiger, M., Hutchinson, J.P., Bingham, R., Ramon, F., Bardera, A., and Martin, J. A Modular, Fully Integrated Ultra-High-Throughput Screening System Based on Confocal Fluorescence Analysis Techniques. (2003), J. Biomol. Screening, 8, 648–659.
- Eggeling, C., Brand, L., Ullmann, D. and Jäger, S. Highly sensitive fluorescence detection technology currently available for HTS. (2003), Drug Discovery Today, Vol. 8, No. 14, 632-641.
- Auer, M., Moore, K.J., Meyer-Almes, F.J., Guenther, R., Pope, A.J. and Stoeckli, K.A. Fluorescence correlation spectroscopy: lead discovery by miniaturized HTS. (1998), Drug Discovery Today, 3, 10, 457-465.
- Pope, A.J., Haupts, U.M. and Moore, K.J. Homogeneous fluorescence readouts for miniaturized high-throughput screening: theory and practice. (1999) Drug Discovery Today, 4, 8, 350-362.
- Haupts, U., Ruediger, M. and Pope, A.J. Macroscopic versus microscopic fluorescence techniques in (ultra)-high-throughput screening. (2000), High-Throughput Screening, 1, 1, 3-9.
- Zhang, J., Chung, T. and Oldenbourg, K. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. (1999), J. Biomol. Screening, 4, 67–73.
Further information
http://www.zinsser-analytic.com/
http://www.evotec-technologies.com
http://www.genomicsolutions.com
http://www.moleculardevices.com




