Basics of image analysis in High Content Screening
Posted: 12 December 2009 | Prof Jeremy Simpson, Professor of Cell Biology and Vasanth Singan, PhD student (Bioinformatics and Computational Biomedicine) University College Dublin | No comments yet
Automated high content screening platforms are capable of producing thousands of images per day. The challenge is to use appropriate analysis methods to extract the maximum amount of biologically-relevant information from these images. In this article we summarise the basic concepts of image analysis and highlight examples of both open-source and commercial software that are available for use with image data sets generated using high-throughput methods.
Automated high content screening platforms are capable of producing thousands of images per day. The challenge is to use appropriate analysis methods to extract the maximum amount of biologically-relevant information from these images. In this article we summarise the basic concepts of image analysis and highlight examples of both open-source and commercial software that are available for use with image data sets generated using high-throughput methods.
Automated high content screening platforms are capable of producing thousands of images per day. The challenge is to use appropriate analysis methods to extract the maximum amount of biologically-relevant information from these images. In this article we summarise the basic concepts of image analysis and highlight examples of both open-source and commercial software that are available for use with image data sets generated using high-throughput methods.
In recent years there has been a trend in both the academic and pharmaceutical arenas towards the production of ever larger and more complex image-based data sets. The automated screening microscopy platforms that generate such data have become increasingly sophisticated and efficient, with the result being that it has become routine to produce thousands or even tens of thousands of high quality images in a single day. This move towards high-throughput biology approaches, and in particular cell-based assays, is one consequence of the successful sequencing of many organisms, providing us with the basic tools to study cells in a truly systematic manner, and ultimately this should enable a more rapid understanding of the entire organism. Traditionally analyses of cell function, or the responses of cells to compounds, have been tested in relatively simple assays, either biochemically-based or using basic fluorescence readings from entire wells of multi-well plates. However it is now increasingly realised that more detailed measurements need to be made from individual cells, and furthermore that it is desirable to analyse multiple parameters in parallel.
Images of cultured cells contain a wealth of information. Using appropriate fluorescent tracers or antibodies it is possible to visualise changes in the spatial distribution or amounts of molecules, in turn providing a read-out of a biochemical process of interest. If experiments are carried out in a time course, or even time-lapse format, temporal information is also gained. The problem therefore is how to analyse such data. Not only are many of the potential phenotypes very subtle and therefore hard to appreciate with the human eye, but accurate conversion of image into numeric data is essential if huge data sets are to be tackled. High content analysis (HCA) has now emerged as a key tool to specifically analyse image data and convert complex morphological parameters extracted from individual cells into a relatively simple numerical output, allowing researchers to rapidly identify cellular phenotypes. HCA is still a comparatively young field, but it is being strongly driven by parallel advances in miniaturisation, robotics, genomics and imaging, with the result being that high content screening (HCS) applications range from basic academic research, through drug discovery to nanotechnologies1. HCS is now widely used in the pharmaceutical industry for cytotoxicity and apoptosis studies, in addition to mainstream drug screening, but its success is only guaranteed if image analysis software is able to deliver truly accurate representation of cellular morphological parameters. In this article we discuss basic image analysis techniques, highlight some existing proprietary and open-source tools for use in HCS, and comment on the current limitations and future developments envisaged for this field.
Choice of HCA strategy
Any cell-based assay needs to be sufficiently robust such that it can be executed over an extended period of time without loss in performance or quality. Similarly the image analysis routines employed must be designed to provide maximum consistency through the life of a screen. HCS experiments are inherently highly parallel, producing large volumes of data, and so the initial choices of software and parameters to be measured are critical. Ideally multiple image features need to be analysed in an automated and systematic manner that minimises human error and bias. The analysis regime must also be sufficiently sensitive to capture all the possible phenotypes expected from the experiment – and this can be a difficult challenge if these are unknown.
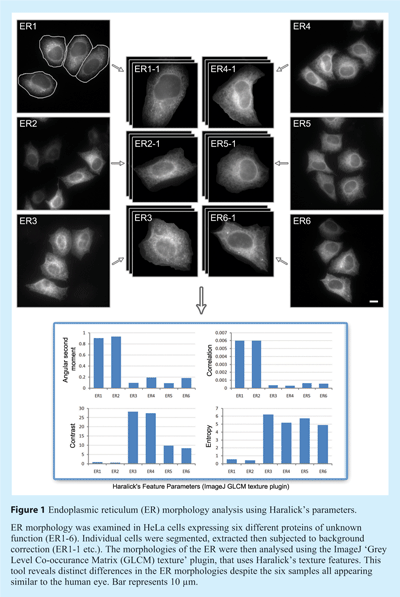
Design of the HCA strategy is very much dependent on the particular assay, and therefore the choice of software employed, whether commercial, open-source, or custom, is critical. Basic assays might include measuring the uptake of a fluorescent ligand into a cell, or the translocation of a molecule between the cytoplasm and nucleus. These events are relatively simple in terms of analysis, and involve standard routines for background subtraction, cell identification and then measurement of a particular area of interest (discussed in more detail below), and as such are all well served by existing software. By contrast, if the assay is more complex, for example analysing changes in subcellular organelle morphology after various treatments, then clearly a greater number of parameters will need to be acquired (see Figure 1). As the organelle of interest become increasingly complex (from a morphological point of view) – for example the endoplasmic reticulum or mitochondria – the challenge of successful and accurate analysis becomes greater. Although a wide variety of analysis software is now available, and many of these have routines that may be usefully applied to these more difficult problems, it may be that a custom solution is still required to detect complex phenotypes of interest. However, the development of such HCA software requires programming knowledge, and is only a realistic option if expertise and resources permit.

Basic routines used in image processing
Successful analysis of HCS data is highly dependent on the quality of the initial images. Automated image acquisition is likely to result in a relatively higher number of poor quality images compared to manual acquisition owing to the inherent nature of autofocus and parallel acquisition. This often leads to many images needing to be filtered or pre-processed before any quantitative information can be extracted. It is therefore advisable that all images are pre-processed to ensure they are of a minimum quality for quantitative analysis. Pre-processing not only enhances images but also saves downstream computational time with respect to final analysis and quantification. Images acquired from cell-based assays need to be channelled through a series of routines in order for the raw image data to be ultimately converted into numeric data. Typically these routines remove background signals, extract individual cells and then identify and quantify morphological features. These are described in greater detail below.
A significant time saving step before any processing is executed is the removal of out-of-focus images and images where there are too few cells for making meaningful quantitative measurements. While the human eye can easily detect out-of-focus images, computer-based recognition of such images is more problematic. Out-of-focus images are blurred, with the fluorescence intensity appearing scattered and at a lower level compared to focused images. One approach to identifying out-of-focus images is to make use of Point Spread Function (PSF). The PSF is derived from the imaging system and the way it responds to the light detected. Critical parameters of the optical system include the numerical aperture of the objectives and the distance of the light source from the detector. The PSF for a particular imaging system can be defined using ‘spread parameters’, with the spread parameters of the PSF of a focused image generally being low, but increasing with defocus. This information can be used to discriminate in- and out-of-focus images, specifically using algorithms designed for the optical system acquiring the images2.
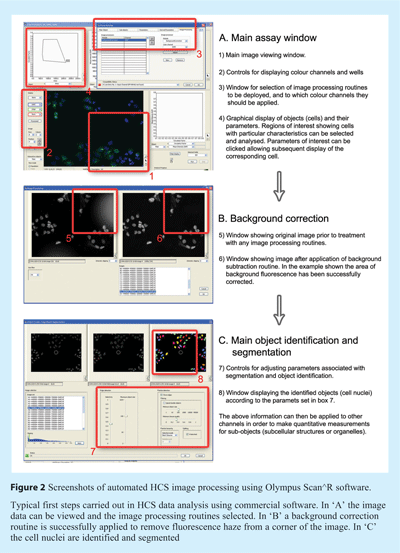
The next issue usually addressed in image analysis is background correction and subtraction. Background fluorescence is a common problem with microscopic acquisitions and can result from wide-spread low-level auto-fluorescence captured by the imaging system, or small pinpoints of intense fluorescence from particles or precipitates (see example in Figure 2). Effective correction of the background largely facilitates the subsequent image segmentation and quantification steps, with the general aim being to reduce the grey levels in the background (for example outside the cells) to zero. One crude approach is to estimate the mean pixel intensity of the background signal and subtract this value from the whole image. This process is ineffective however if the background is uneven, and often results in loss of data and so must be used cautiously. Many algorithms estimate the background value based on the illumination, detector gain and offset of the imaging system, and compare this with the acquired image, thereby determining the correction needed. An alternative method uses histogram-based background correction routines, although this method is more applicable for use with images containing sub-confluent concentrations of cells. Such routines work by measuring the distribution of pixel intensities across the entire image, allowing a fitting parabola to be drawn through the maximum values in the histogram, and the background estimated from this information. Another technique used in some commercial software utilises the so-called ‘rolling ball algorithm’. With this technique a local background value is determined by averaging values over a large ball around the pixel and this value is subtracted from the image. One advantage of this technique is that the user has control over the size of ball used, and therefore it can easily be adapted to different assays and image types.

The next challenge with processing microscopy images of cells is segmentation. This is the process of identifying, partitioning and extracting individual cells in the field of view for subsequent analysis. Segmentation can be done on a pixel-by-pixel basis assigning each pixel into one of the segments based on various features including the pixel intensity, texture, and colour. Various methods and algorithms exist to perform image segmentation and open-source toolkits like ITK3 can be used for registration and segmentation. Segmentation can be broadly classified into region-based and boundary-based methods. Region-based segmentation methods group similar intensity pixels into common regions. Thresholding is one such commonly used region-based segmentation technique and uses simple Boolean classification of pixels and works well with uniform grey levels of objects. Other region-based segmentation algorithms like gradient-based algorithms and watershed algorithms exist for segmentation. This latter technique is particularly powerful and is used widely in commercial software. It works by searching for areas of lower pixel intensity between areas of high intensity, effectively making a series of valleys and hills. The lowest points of the valleys effectively mark the cell edges. Boundary-based segmentation methods work by looking for areas of sudden change in intensity between adjacent pixels. For example, Laplacian Image thresholding is one such method often used. The Canny Edge detector is another efficient tool for noise-sensitive data and uses a multistage edge detection algorithm.
Once individual cells have been identified, phenotypic and subcellular information can begin to be classified based on a set of features extracted from each of the segmented objects (usually individual cells) and their associated sub-objects (usually subcellular organelles). The number of features that can be observed is potentially limitless and is constrained only by image processing capacity. Based on the particular experiment, biological conditions, cell lines, etc., appropriate features can be quantified and characterised. Broadly speaking, these features are based on geometry (for example perimeter of object, size, and circularity factor), pixel intensity, pixel distribution, and texture. One of the most commonly used features is based on Haralick’s co-occurrence features which use co-occurrence distribution of pixel values to generate information about the texture of objects4 (see Figure 1). Several instances of its use in cell phenotype classification have been reported in the literature.5,6 Altogether the potentially hundred or so common features that can be extracted provide a series of quantitative measurements that relate to the appearance of the cell, thus marking the completion of the analysis part of work.
The final step in the analysis pipeline is classification of the objects and associated sub-objects. Automation of phenotypic and morphological classification is critical in high throughput experiments, and once again robust tools are required to ensure that classification is carried out in a meaningful and statistically significant manner. Classifiers like Bayes use a priori probabilities of class, and estimate the probability of each extracted feature belonging to a particular class based on the probability density function of the class. Machine learning algorithms for classification can be supervised (training data set) or unsupervised (model-based). Supervised learning involves manual classification of features from a subset of data to train the system. Supervised learning algorithms like k-nearest neighbours, support vector machines, and naive Bayesian classifiers are commonly used in biological applications, and many good examples of their use have been reported7 and are reviewed8. Unsupervised learning involves clustering of objects based on the maximum variance and maximum correlation to group objects. Based on the application, prior knowledge and computational cost involved, the choice of using supervised or unsupervised learning algorithms is also important. Unsupervised learning can result in a large number of unknown classes and is often more time consuming. Supervised learning requires prudent selection of the training set and if the set is not exhaustive, important features might be undetected. While there are advantages and disadvantages for both the methods, the choice depends on the data and resources available.
Programming environments
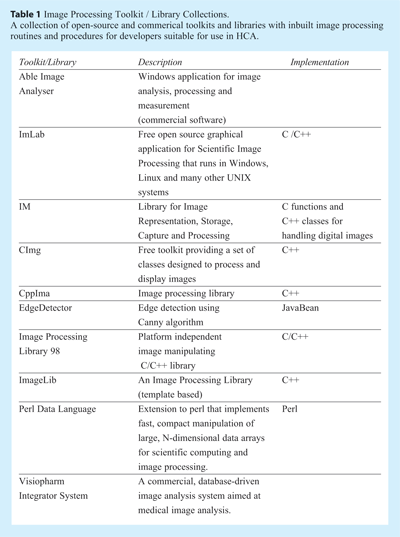
As described above, the analysis pipeline for HCS images can be complex, and if the assay is not particularly suited to analysis by commercial software or routines then the development of a custom solution may be required. Although many of the routines needed for image processing are generic, the development of customised software is not trivial, and strong computational expertise is a necessity. There are rich programming languages that can help developers in designing image analysis software. Programming Languages including Java and Matlab are well established, with both providing routines for image processing applications, visualisation and algorithm development. Open-source programming languages like Java provide extensive packages with modules for image processing. The programming language C / C++ can also be used for developing efficient image processing algorithms. Table 1 shows some examples of toolkits and image processing libraries (both open-source and commercial) in various languages that can be used by developers to fully take control of their HCA needs.
Existing open-source and commercial software for HCA
The growing use of HCS in biology has spawned a massive increase in analysis software. Although detailed comparisons of all HCA software is beyond the scope of this article a selection of software, both open-source and commercial, which are familiar to this laboratory are briefly discussed below.
1. ImageJ
ImageJ (http://rsbweb.nih.gov/ij/index.html) is a Java-based image processing platform that enables display, editing, analysis and processing of digital images in a variety of formats. Multithreaded processing in ImageJ enhances speed of operations as they are performed in parallel. It is an open-source tool that comes with a suite of plugins for image processing and researchers are encouraged to contribute to and download plugins according to their needs. It supports standard image processing functions such as contrast manipulation, sharpening, smoothing, edge detection and median filtering. However, ImageJ lacks the capability to automatically analyse very large data sets, and so ‘wrapper’ programs or applications might be needed if this platform is to be applied to HCS data. Nevertheless, the open-source development environment encourages developers worldwide to contribute and use the plugins and it has become a powerful means for exchanging image processing routines. Below are a few examples of plugins that have been shared by developers and that are appropriate to HCA needs.
- Circularity – an extended version of ImageJ’s Measure command that calculates object circularity
- Cell Counter – a plugin for counting cells and has features to add different counter types
- Microscope Scale – a plugin for calibrating images spatially, using hard-coded arrays of magnifications, calibration values and length units
- Colocalisation – a plugin to create colocalisation points of two 8-bit images
- Granulometry – a plugin to extract size distribution from binary images
- Texture Analysis – a plugin to compute Haralick’s texture parameters.

2. CellProfiler
CellProfiler is free cell image analysis software developed at the Broad Institute, and is designed for specific use with multidimensional data from high-throughput experiments9. It also contains a supervised machine learning system that can be trained to recognise complicated and subtle phenotypes, enabling automatic scoring of millions of cells. It is designed with a modular approach using gating of individual cells to score complex phenotypes and hence classify hits. CellProfiler allows users to build their own pipeline of individual modules that suit their particular assay. This gives greater flexibility for the users in terms of choosing appropriate modules and avoiding unnecessary ones. CellProfiler Analyst builds upon CellProfiler and is designed for high-end exploration and analysis of measured features from high-throughput image-based screens10.
3. DetecTiff
DetecTiff is a newly reported image analysis software that can be used for automated object recognition and quantification of digital images11. Written in the LabView environment from National Instruments, it uses template-based processing for quantitative analysis, with algorithms for structure recognition based on intensity thresholding and size-dependent particle filtering. DetecTiff enables processing of multiple detection channels and provides functions for template organisation and fast interpretation of acquired data. DetecTiff allows users to customise and set parameters that can then be used for fully automated analysis. DetecTiff has been shown to produce quantitative results comparable to CellProfiler and appears to be efficient at processing large data sets from screens.
4. BioImageXD
BioImageXD is open-source software for image analysis and processing. It is designed to work with single or multi-channel 2D, 3D and 4D (time series) image data12. BioImageXD has features for realistic 3D image rendering and provides users with various viewing modes including slices and orthorgraphic sections. It comes with a set of basic image processing routines and also 3D segmentation and analysis features. BioImageXD is written in Python and C++ and uses the ITK toolkit for segmentation and image processing tasks. It also has a colocalisation analysis routine for analysis of signal intensities in 3D stacks.
5. Scan^R Analysis
Scan^R Analysis is a proprietary analysis software from Olympus Soft Imaging Solutions, and although it is primarily designed for use with image data acquired on Olympus Scan^R automated screening microscopes, it can handle large data sets from other high content systems. It has a set of modules for performing analysis, quantification and navigation through the results, and these can be run during analysis or in ‘off-line’ mode. The various image processing and quantification procedures can be defined as an assay and stitched together to perform sequentially (see Figure 2). The main interface is in the form of histograms for easy selection of objects with features of interest, highly similar to software used to analyse flow cytometery data. The most recent release of the software also has inbuilt procedures for particle tracking in time-lapse data, which although is limited in terms of throughput, highlights the trend towards performing time-resolved assays in living cells.
6. Cellenger
Cellenger is a commercial software from Definiens specifically designed for HCS applications. It is composed of a set of workflow tools and is capable of working on multiple platforms. Its modular environment allows users to select analysis routines as needed and it is capable of working with large data sets.
Limitations and future developments
Within a relatively short period of time cell-based assays and their analysis have become an important tool for biologists seeking greater insight into cell health and function. While the potential of this approach is clear, its further use faces a number of challenges. From the time since Cellomics pioneered the first automated screening platform many other manufacturers have now developed powerful HCS systems. The pace of these events has been so rapid that standards and formats for images and metadata have not yet been truly standardised. Due to the volumes of HCS data produced and the fact that images may need to be analysed by different software if the maximum amount of information is to be extracted, improved standardisation is essential. A common platform and controlled vocabulary for easy exchange and seamless integration of various analysis tools would also be welcome. Further improvements in image analysis software are also expected to enhance the information gained from HCS regimes, but parallel increases in computer processing power are needed if data analysis and retrieval are to remain efficient. Finally it is worth noting that the future of cell-based assays will also see more use of experiments carried out in living cells in time-lapse format. While this will undoubtedly deepen our biological knowledge, it will also provide new challenges to data storage and analysis.
References
- Bickle M (2008). High-content screening: a new primary screening tool? IDrugs 11:822-826.
- Wu Q, Merchant F and Castleman KR (2008). Microscope Image Processing. Pub. Academic Press.
- Yoo TS, Ackerman MJ, Lorensen WE, Schroeder W, Chalana V, Aylward S, Metaxes D and Whitaker R (2002). Engineering and algorithm design for an image processing API: a technical report on ITK – The Insight Toolkit. In Proceedings of Medicine Meets Virtual Reality, J. Westwood, ed., IOS Press Amsterdam pp 586-592.
- Haralick RM (1979). Statistical and structural approaches to texture. Proceeding of the Institute of Electrical and Electronics Engineers (IEEE) 67:786-804.
- Wang J, Zhou X, Bradley PL, Chang S, Perrimon N and Wong STC (2008). Cellular phenotype recognition for high-content RNA interference genome-wide screening. J. Biomol. Screen. 13:29-39.
- Tsai YS, Chung IF, Simpson JC, Lee MI, Hsiung CC, Chiu TY, Kao LS, Chiu TC, Lin CT, Lin WC, Liang SF and Lin CC (2008). Automated recognition system to classify subcellular protein localizations in images of different cell lines acquired by different imaging systems. Microsc. Res. Tech. 71:305-314.
- Conrad C, Erfle H, Warnat P, Daigle N, Lorch T, Ellenberg J, Pepperkok R and Eils R (2004). Automatic identification of subcellular phenotypes on human cell arrays. Genome Res. 14:1130-1136.
- Wollman R and Stuurman N (2007). High throughput microscopy: from raw images to discoveries. J. Cell Sci. 120:3715-3722.
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, Golland P and Sabatini DM (2006). CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7:R100.
- Jones TR, Kang IH, Wheeler DB, Lindquist RA, Papallo A, Sabatini DM, Golland P and Carpenter AE (2008). CellProfiler Analyst: data exploration and analysis software for complex image-based screens. BMC Bioinformatics 9:482.
- Gilbert DF, Meinhof T, Pepperkok R and Runz H (2009). DetecTiff: A novel image analysis routine for high-content screening microscopy. J. Biomol. Scr. 14:944-955.
- Kankaanpää P, Pahajoki K, Marjomäki, V, Heino J and White D (2006). BioImageXD – new open source free software for the processing, Analysis and visualization of multidimensional microscopic images. Microscopy Today, 14(3):12-16.




