Proteomics research advanced by use of AI
Posted: 30 May 2019 | European Pharmaceutical Review | No comments yet
Researchers succeed in using machine learning to make the mass analysis of proteins faster and almost error-free.
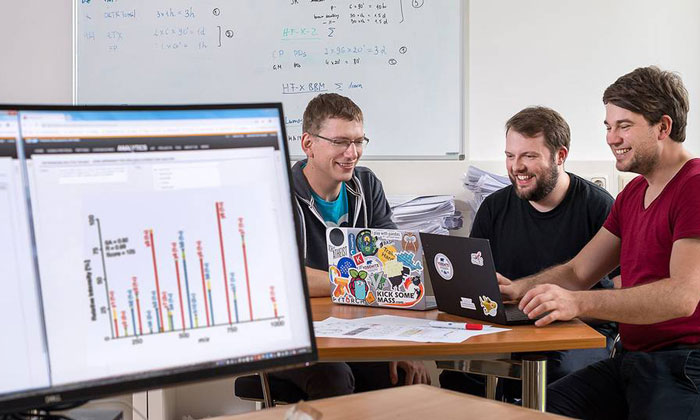

The authors Mathias Wilhelm, Tobias Schmidt and Siegfried Gessulat (credit: A. Eckert /TUM).
The researchers (from the Technical University of Munich and led by bioinformatics scientist Mathias Wilhelm and biochemist Bernhard Küster, Professor of Proteomics and Bioanalytics at the Technical University of Munich) successfully used proteomic data to train a neural network to recognise proteins more quickly and with almost no errors.
Currently, certain proteins are not recognised or are recognised incorrectly with mass spectrometers due to them not measuring proteins directly, which is “a serious problem,” said Bernhard Küster. The neural network developed by the TUM team uses all the information of the spectra for the process of identification. “We miss fewer proteins and make 100 times fewer mistakes,” he revealed.
The AI software (named ‘Prosit’) is “applicable to all organisms in the world, even if their proteomes have never been examined before,” explained Mathias Wilhelm. “This enables research which was previously inconceivable.”
With the help of 100 million mass spectra, the algorithm has been so extensively trained that it can be used for all common mass spectrometers without any additional training. “Our system is the global leader in this field,” added Küster.
With Prosit, it will be possible to develop even more powerful instruments like this in the future. Researchers and physicians will also be better and faster able to search for biomarkers in patients’ blood or urine, or monitor therapies for their effectiveness.
The researchers also have high hopes for fundamental research: “The method can be used to track down new regulatory mechanisms in cells,” said Küster. “We hope to gain a considerable amount of knowledge here, which, in the medium and long term, will be reflected in the treatment of diseases suffered by humans, animals and plants.”
This research was published in Nature Methods.
Related topics
Artificial Intelligence, Mass Spectrometry, Proteins, Research & Development (R&D)