Small non-coding RNAs as therapeutics
Posted: 20 March 2009 | Dr Kevin V. Morris, Assistant Professor, Department of Molecular and Experimental Medicine, The Scripps Research Institute and Peter Hawkins, Graduate student, Kellogg School of Science and Technology, The Scripps Research Institute | No comments yet
For years biologists have worked to develop alternatives to traditional therapeutics. These efforts, in areas such as stem cell based and gene therapies, have received much fanfare in popular media outlets, raising expectations among the general public. This excitement may have contributed to the hasty progression of early gene therapy trials, which tragically led to several deaths. Despite early failures in the development of gene therapies, work in this field has continued, and the promise of life saving treatments remains.
For years biologists have worked to develop alternatives to traditional therapeutics. These efforts, in areas such as stem cell based and gene therapies, have received much fanfare in popular media outlets, raising expectations among the general public. This excitement may have contributed to the hasty progression of early gene therapy trials, which tragically led to several deaths. Despite early failures in the development of gene therapies, work in this field has continued, and the promise of life saving treatments remains.
For years biologists have worked to develop alternatives to traditional therapeutics. These efforts, in areas such as stem cell based and gene therapies, have received much fanfare in popular media outlets, raising expectations among the general public. This excitement may have contributed to the hasty progression of early gene therapy trials, which tragically led to several deaths. Despite early failures in the development of gene therapies, work in this field has continued, and the promise of life saving treatments remains.
Early gene therapy strategies consisted primarily of the introduction of a functional gene to compensate for a mutated or otherwise nonfunctional endogenous allele. While this approach is appropriate for many serious conditions, such as hemophilia and X-linked severe combined immunodeficiency (X-SCID), genetic diseases caused by the inappropriate upregulation of gene expression cannot be addressed by this method. An alternative approach designed to treat diseases caused by upregulated gene expression is antisense therapy, a process whereby an RNA molecule is introduced which hybridises to the mRNA of the upregulated gene in question, blocking its translation1.
In addition to being a monumental finding for basic research, the discovery of RNA interference (RNAi), led to immediate excitement regarding the application of this technology for therapeutic benefit, specifically as a novel mode of gene therapy. This review will provide a basis for description of the biology of RNAi along with a discussion of the current state of RNAi based therapy development and various obstacles facing the field.
RNA interference: Post Transcriptional Gene Silencing
RNA interference (RNAi) is the process by which small double-stranded RNA molecules (dsRNAs) induce homology dependent inhibition of gene expression2-4. This phenomenon is known as quelling in the filamentous fungus Neurospora crassa5-7 and was originally described in plants where it was termed co-suppression7. Since these early observations of RNAi, it has became apparent that RNAi functions to suppress gene expression by two distinct pathways: transcriptional gene silencing (TGS) and post-transcriptional gene silencing (PTGS)8-9. The better characterised molecular pathway, PTGS, occurs when small dsRNAs are targeted to a gene transcript (mRNA). The dsRNA trigger for this process can be of endogenous (micro RNA: miRNA) or exogenous (small-interfering RNA: siRNA) origin. In human cells, miRNAs are transcribed as hairpins, termed primary RNAs (pri-RNAs). Pri-RNAs are processed in the nucleus by the RNAse III enzyme Drosha to form a slightly shortened hairpin, or pre-miRNA. Pre-mRNAs are exported from the nucleus and processed into mature miRNAs by a second RNAse III enzyme, Dicer, which cleaves the loop of the hairpin, leaving a dsRNA ~21-23 nucleotides long. Synthetic double stranded RNAs (siRNAs) resemble mature miRNAs in structure and enter the pathway at this point. The guide strand of the small dsRNA is incorporated in the Argonaute 2 (Ago2) and Dicer containing RNA induced silencing complex (RISC), which is the effector complex for the PTGS pathway. Loaded RISC can function to inhibit gene expression by several mechanisms. These include inhibiting expression through interactions with the 3’UTR or by directly targeting the homologous genes mRNA for degradation or translational inhibition10, which activities appear to take place in cytoplasmic P-bodies11. Message targeted miRNAs may also function to repress translation by interactions between RISC and the ribosome12. PTGS has been shown to operate in both the cytoplasm and nucleus of human cells13-15.
Despite its relatively recent discovery, RNAi is already being investigated as an approach to treat a wide spectrum of diseases. According to The Journal of Gene Medicine Clinical Trial site, RNAi is being investigated in the clinic to combat the effects of HIV, RSV, macular degeneration, chronic hepatitis B, and other diseases (http://www.wiley.co.uk/ genetherapy/clinical/).
RNAi: Transcriptional Gene Silencing
The majority of research in the field of RNAi has focused on siRNA and/or miRNA directed PTGS of cellular mRNAs. However, in human cells small RNAs can also function to suppress gene expression by inducing epigenetic modifications at targeted gene promoters, resulting in transcriptional gene silencing16. Epigenetics is the study of meiotically and mitotically heritable changes in gene expression which are not coded for in the DNA17,18. Common types of epigenetic changes include silencing by CpG methylation, and silencing and activation by various histone modifications. Small RNA mediated TGS was first observed when doubly transformed tobacco plants exhibited a suppressed phenotype of a transgene that correlated with DNA methylation at the transgene promoter19. While TGS is a well established regulatory mechanism in plants and yeast, work in human cells has lagged behind. Thanks to many recent published reports, however, a model for TGS in human cells has begun to take shape.
In this model, the generation and activity of small RNA molecules requires Argonaute 1(Ago1)20,21. Target recognition occurs as the antisense strand of the small RNA binds to a complementary promoter-associated RNA (pRNA)22,23 (see Figure 1). Interestingly, several recent reports have shown that transcription of promoter regions is much more common that previously realised24-26. Upon exposure to the small non-coding RNA, the targeted promoter exhibits higher levels of the silent state histone methyl marks H3 lysine-9 di-methylation (H3K9me2) and histone H3 lysine-27 tri-methylation (H3K27me3)22,27-29 (see Figure 1). These silent state chromatin marks have been observed not only at the targeted locus but also downstream, 3′ of the target site, providing a possible mechanism by which silent state histone methyl marks may extend to affect neighboring genes or intergenic regions27,30. A role for DNA methylation in this process remains uncertain, as some targeted promoters have been observed to exhibit increased CpG methylation31, while others have not32.
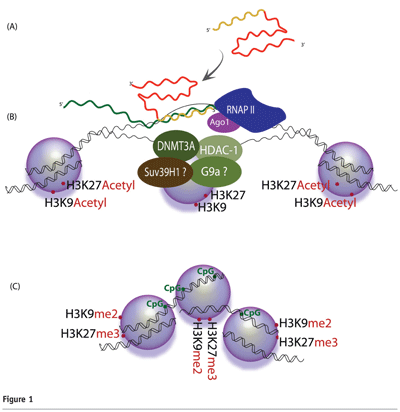

Protein factors responsible for these enzymatic processes constitute a partially characterised transcriptional silencing complex (TSC). Recent work indicates that this putative TSC may contain DNMT3a27,33, Ago-128,29, HDAC-133,34, and histone methyltransferases such as enhancer of zeste 2 (Ezh2) and/or G9a22,35. In some instances Argonaute 2 (Ago2) has been shown to be required for TGS28, and recently published work indicates that the varying incorporation of Ago1 and Ago2 may depend on the level of complementarities between the small RNA and its target36.
TGS is an attractive avenue for the development of gene therapies. In addition to levels of efficacy and specificity similar to PTGS, TGS is advantageous in that it can induce a long-term, heritable change in gene expression. Long-term inhibition of transcription is a result of the epigenetic mechanisms involved, which are inherently stable and longer lasting than the effects induced by PTGS. While there are no clinical trials for small RNA mediated TGS based therapeutics, research towards such a goal is taking place. TGS is currently being investigated as a basis for treating a wide range of diseases from cancer to HIV.
Bidirectional transcription as an endogenous TGS mechanism
Recently, work in the field of TGS has turned towards efforts to identify and describe endogenous RNA triggers in human cells. One exciting finding in this area is the discovery that RNA-mediated TGS appears to play an important role in early transcriptional silencing events which lead to human X-inactivation37. In addition, two newly published reports have identified the first examples of endogenously expressed promoter targeted miRNAs which mediate this process20,36. It has also been shown that some non-coding, antisense transcripts may direct TGS to their respective sense promoters38, and that targeting these antisense transcripts can lead to activation of sense transcription23,31. Recent genome wide studies indicate that nonrandom antisense transcription is widespread in the human genome, and may be a prevalent mechanism of transcriptional regulation39,40.
The discovery that transcription of a coding mRNA can be in part regulated by the relative abundance of its antisense non-coding RNA has many implications in basic science as well as applied research. Besides the fact that this finding describes a possible endogenous mechanism of transcriptional regulation, the role that antisense transcripts may play in gene regulation may provide additional insight into novel possible ways that miRNAs may function. In human cells, many miRNAs target the 3’UTR of a given transcript. Interestingly, during our database screens for bidirectional transcripts in various genes, many antisense RNAs overlap with these regions, and indeed we have identified some known miRNA sites which overlap with areas of genes which are bidirectionally transcribed. While little is known about such a mechanism, one could envision a scenario in which miRNAs targeted to bidirectionally transcribed gene regions could differentially regulate the levels of sense and antisense transcripts in such a way to not only post-transcriptionally, but also transcriptionally regulate gene expression.
In addition to the importance of these observations in the realm of basic research, they may open new avenues for the development of a novel class of gene therapy. Gene regulation by bidirectional transcription may prove to be an additional system that can be manipulated for therapeutic benefit. For example, many cancers are believed to be caused by the misregulation of certain genes, such as the inappropriate upregulation of oncogenes or the inappropriate downregulation of tumour suppressor genes. It may be possible to fine tune the expression of such genes by selectively targeting sense or antisense transcripts to ultimately increase or decrease their mRNA transcription. Such fine tuning could be accomplished theoretically with small antisense non-coding RNAs, siRNAs, or genetic therapy based vector systems. Interestingly, database investigation of many well-characterised oncogenes and tumour suppressor genes indicates that many may be bidirectionally transcribed.
Obstacles
From the onset of investigations of gene therapy, specific delivery of the therapy to target cells has proven to be a significant obstacle. In fact, the retroviral vector used in early X-SCID trials was ultimately blamed for causing leukemia which led to the tragic deaths of several trial subjects. The cause of these problems stems from integration of proviral DNA into promoter elements. Such retroviral integrations can cause misregulation of gene expression, ultimately contributing to oncogenesis. Since these early failures in targeted delivery, much progress has been made in developing effective and safe vectors for therapeutic delivery. Current clinical trials are utilising many vectors, most predominately adenoviruses and lentiviruses. In addition to viral vectors, progress has been made using alternative delivery methods. Of such include the development of nanoparticle based delivery systems and the injection of naked nucleic acid41.
Since the discovery of RNAi, many observations have been made regarding the possibility and reality of “off-target effects.” Off-target effects can occur during RNAi applications as mi- or siRNAs can recognise and bind sequence elements in addition to their desired target. In addition to this important negative side-effect of RNAi treatments, recent work in the field of TGS has uncovered another possible type of off-target effect. It has been shown that epigenetic modifications targeted to gene promoters by small RNAs can spread down-stream (5′-3′ from the target site). It has also recently been shown that small RNAs targeted to coding regions can also direct epigenetic modifications to their respective regions in the genomic DNA16. While such a phenomenon has not been observed yet, it may be possible that a siRNA introduced into a cell as part of a therapeutic may, in addition to recognising its designed target, direct epigenetic modifications to the corresponding genomic locus. These epigenetic modifications could then spread downstream, and, if the target site is located near the 3′ end of a coding region, could lead to the remodeling of an adjacent gene promoter. Such a scenario should be considered when investigators plan to utilise the RNAi pathway for research or medicine.
While the use of the TGS pathway of RNAi for gene therapy applications holds some advantages over the PTGS pathway, there are some additional considerations. These potential problems stem from the perceived advantage of TGS, specifically that long-term, heritable gene silencing can be achieved. Recently published work has shown that epigenetic changes induced by environmental stimuli, in the case of the study famine, can be passed on to subsequent offspring42. Several oncogenes which could in theory be targeted by TGS to prevent tumourogenesis in adults are necessary during development. If these genes are epigenetically silenced in a reproducing adult, these changes could be passed on to offspring, leading to developmental complications. In light of these observations, those pursuing future clinical applications of TGS should carefully consider the reproductive status of potential subjects or patients.
Conclusion
RNA interference is an important regulatory mechanism whereby gene expression can be silenced both transcriptionally and post-transcriptionally. Besides the obvious importance of this discovery for basic research, RNAi holds much promise for applied medical research as well. Manipulation of gene expression to counteract disease is a promising area of medical research, despite early shortcomings and an inability to meet early, and possibly unreasonable, public expectations. While significant obstacles remain, progress towards effective RNAi-based gene therapies is ongoing, and this active area of research may prove to be the source of future therapeutics.
Acknowledgements
This project is funded by R01 HL083473-02 to KVM. I thank Paula J. Morris at Seainsite http://seainsite.com/index.html for the generation of figures.
References
- Stein, C. A. and J. S. Cohen (1989). “Antisense compounds: potential role in cancer therapy.” Important Adv Oncol: 79-97.
- Montgomery, M. K., S. Xu, A. Fire. (1998). “RNA as a target of double-stranded RNA-mediated genetic interference in Caenorhabditis elegans.” Proceedings of the National Academy of Sciences 95: 15502-15507.
- Nishikura, K. (2001). “A short primer on RNAi: RNA-directed RNA polymerase acts as a key catalyst.” Cell 107: 415-418.
- Sharp, P. A. (2001). “RNA interference.” Genes and Development 15: 485-490.
- Fire, A., S. Xu, M.K. Montgomery, S.A. Kostas, S.E. Driver, C.C. Mello. (1998). “Potent and specific genetic interference by double stranded RNA in Caenorhabditis elegans.” Nature 391: 806-811.
- Hutvagner, G. and P. D. Zamore (2002). “RNAi: nature abhors a double-strand.” Curr Opin Genet Dev 12(2): 225-32.
- Tijsterman, M., R. F. Ketting, et al. (2002). “The genetics of RNA silencing.” Annu Rev Genet 36: 489-519.
- Sijen, T., I. Vign, A. Rebocho, R. Blokland, D. Roelofs, J. Mol, and J. Kooter. (2001). “Transcriptional and posttranscriptional gene silencing are mechansitically related.” Current Biology 11: 436-440.
- Pal-Bhadra, M., U. Bhadra, J. A. Birchler. (2002). “RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in drosophila.” Molecular Cell 9: 315-327.
- Leung, A. K. and P. A. Sharp (2006). “Function and localization of microRNAs in mammalian cells.” Cold Spring Harb Symp Quant Biol 71: 29-38.
- Liu, J., M. A. Valencia-Sanchez, et al. (2005). “MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies.” Nat Cell Biol 7(7): 719-23.
- Chendrimada, T. P., R. I. Gregory, et al. (2005). “TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing.” Nature 436(7051): 740-4.
- Langlois, M. A., C. Boniface, et al. (2005). “Cytoplasmic and nuclear retained DMPK mRNAs are targets for RNA interference in myotonic dystrophy cells.” J Biol Chem 280(17): 16949-54.
- Robb, G. B., K. M. Brown, et al. (2005). “Specific and potent RNAi in the nucleus of human cells.” Nat Struct Mol Biol 12(2): 133-7.
- Weinmann, L., J. Hock, et al. (2009). “Importin 8 Is a Gene Silencing Factor that Targets Argonaute Proteins to Distinct mRNAs.” Cell.
- Hawkins, P. G. and K. V. Morris (2008). “RNA and transcriptional modulation of gene expression.” Cell Cycle 7(5): 602-7.
- Jackson, A. L., S. R. Bartz, et al. (2003). “Expression profiling reveals off-target gene regulation by RNAi.” Nat Biotechnol 21(6): 635-7.
- Egger, G., G. Liang, et al. (2004). “Epigenetics in human disease and prospects for epigenetic therapy.” Nature 429(6990): 457-63.
- Matzke, M. A., M. Primig, J. Trnovsky, and A.J.M. Matzke. (1989). “Reversible methylation and inactivation of marker genes in sequentially transformed tobacco plants.” The EMBO Journal 8: 643-649.
- Kim, D. H., P. Saetrom, et al. (2008). “MicroRNA-directed transcriptional gene silencing in mammalian cells.” Proc Natl Acad Sci U S A 105(42): 16230-5.
- Janowski, B. A., K. E. Huffman, et al. (2006). “Involvement of AGO1 and AGO2 in mammalian transcriptional silencing.” Nat Struct Mol Biol.
- Han, J., D. Kim, and K.V. Morris (2007). “Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells.” PNAS 104(30).
- Schwartz, J. C., S. T. Younger, et al. (2008). “Antisense transcripts are targets for activating small RNAs.” Nat Struct Mol Biol.
- Core, L. J., J. J. Waterfall, et al. (2008). “Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters.” Science 322(5909): 1845-8.
- Preker, P., J. Nielsen, et al. (2008). “RNA exosome depletion reveals transcription upstream of active human promoters.” Science 322(5909): 1851-4.
- Seila, A. C., J. M. Calabrese, et al. (2008). “Divergent transcription from active promoters.” Science 322(5909): 1849-51.
- Weinberg, M. S., L.M. Villeneuve, A. Ehsani, M. Amarzguioui, L. Aagaard, Z. Chen, A.D. Riggs, J.J. Rossi, and K.V. Morris. (2005). “The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells.” RNA 12(2).
- Janowski, B. A., S. T. Younger, et al. (2007). “Activating gene expression in mammalian cells with promoter-targeted duplex RNAs.” Nat Chem Biol.
- Kim, D. H., L. M. Villeneuve, et al. (2006). “Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells.” Nat Struct Mol Biol 13(9): 793-7.
- Morris, K. V. (2005). “siRNA-mediated transcriptional gene silencing: the potential mechanism and a possible role in the histone code.” Cell Mol Life Sci 62(24): 3057-66.
- Morris, K. V., S. Santoso, et al. (2008). “Bidirectional transcription directs both transcriptional gene activation and suppression in human cells.” PLoS Genet 4(11): e1000258.
- Ting, A. H., K. E. Schuebel, et al. (2005). “Short double-stranded RNA induces transcriptional gene silencing in human cancer cells in the absence of DNA methylation.” Nat Genet 37(8): 906-10.
- Turner, A. M., J. De La Cruz, et al. (2008). “Mobilization-competent Lentiviral Vector-mediated Sustained Transcriptional Modulation of HIV-1 Expression.” Mol Ther.
- Suzuki, K., T. Juelich, et al. (2008). “Closed chromatin architecture is induced by an RNA duplex targeting the HIV-1 promoter region.” J Biol Chem.
- Vire, E., C. Brenner, et al. (2005). “The Polycomb group protein EZH2 directly controls DNA methylation.” Nature.
- Gonzalez, S., D. G. Pisano, et al. (2008). “Mechanistic principles of chromatin remodeling guided by siRNAs and miRNAs.” Cell Cycle 7(16): 2601-8.
- Ogawa, Y., B. K. Sun, et al. (2008). “Intersection of the RNA interference and X-inactivation pathways.” Science 320(5881): 1336-41.
- Yu, W., D. Gius, P. Onyango, K. Muldoon-Jacobs, J. Karp, A.P. Feinberg, H. Cui. (2008). “Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA.” Nature 451(10 January): 202-206.
- He, Y., B. Vogelstein, et al. (2008). “The antisense transcriptomes of human cells.” Science 322(5909): 1855-7.
- Guttman, M., I. Amit, et al. (2009). “Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals.” Nature.
- Compagno, D., C. Merle, et al. (2007). “SIRNA-directed in vivo silencing of androgen receptor inhibits the growth of castration-resistant prostate carcinomas.” PLoS ONE 2(10): e1006.
- Heijmans, B. T., E. W. Tobi, et al. (2008). “Persistent epigenetic differences associated with prenatal exposure to famine in humans.” Proc Natl Acad Sci U S A 105(44): 17046-9.