Size does matter: macro ncRNAs and the regulation of imprinted gene clusters
Posted: 23 January 2008 | Denise P. Barlow, Stefan H. Stricker and Paulina Latos, CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences | No comments yet
The availability of the human and the mouse sequence has allowed genome-wide analysis of transcription to produce ‘transcriptomes’ that list all RNA transcripts in specific cell types or tissues. These studies have identified a surprisingly large number of ncRNAs that were not recognised by gene annotation programs applied to the genomic sequence. The earliest mouse transcriptome based on sequence annotation of full-length cDNA clones demonstrated that more than 70% of mapped cDNAs arose from non-coding transcripts and 15% of all transcripts formed sense/antisense pairs1. This surprising finding has been confirmed by genomic tiling arrays that allow whole chromosomes or genomes to be simultaneously analysed at a high resolution1-6 and showed that ncRNA transcripts constitutes the bulk of the mammalian transcriptome. To date, studies that quantify non-coding transcription have been performed in most model organisms7. Non-coding transcription is more prominent in higher eukaryotes, indicating that higher genome complexity made it necessary to make use of an additional layer of gene regulation. It should be noted that a recent yeast study showed that some antisense transcription might be an artefact arising from spurious synthesis of second-strand cDNA during reverse transcription reactions8. This indicates the amount of antisense non-coding transcription identified using strategies that did not include Actinomycin D may have been overestimated.
The availability of the human and the mouse sequence has allowed genome-wide analysis of transcription to produce 'transcriptomes' that list all RNA transcripts in specific cell types or tissues. These studies have identified a surprisingly large number of ncRNAs that were not recognised by gene annotation programs applied to the genomic sequence. The earliest mouse transcriptome based on sequence annotation of full-length cDNA clones demonstrated that more than 70% of mapped cDNAs arose from non-coding transcripts and 15% of all transcripts formed sense/antisense pairs1. This surprising finding has been confirmed by genomic tiling arrays that allow whole chromosomes or genomes to be simultaneously analysed at a high resolution1-6 and showed that ncRNA transcripts constitutes the bulk of the mammalian transcriptome. To date, studies that quantify non-coding transcription have been performed in most model organisms7. Non-coding transcription is more prominent in higher eukaryotes, indicating that higher genome complexity made it necessary to make use of an additional layer of gene regulation. It should be noted that a recent yeast study showed that some antisense transcription might be an artefact arising from spurious synthesis of second-strand cDNA during reverse transcription reactions8. This indicates the amount of antisense non-coding transcription identified using strategies that did not include Actinomycin D may have been overestimated.
The availability of the human and the mouse sequence has allowed genome-wide analysis of transcription to produce ‘transcriptomes’ that list all RNA transcripts in specific cell types or tissues. These studies have identified a surprisingly large number of ncRNAs that were not recognised by gene annotation programs applied to the genomic sequence. The earliest mouse transcriptome based on sequence annotation of full-length cDNA clones demonstrated that more than 70% of mapped cDNAs arose from non-coding transcripts and 15% of all transcripts formed sense/antisense pairs1. This surprising finding has been confirmed by genomic tiling arrays that allow whole chromosomes or genomes to be simultaneously analysed at a high resolution1-6 and showed that ncRNA transcripts constitutes the bulk of the mammalian transcriptome. To date, studies that quantify non-coding transcription have been performed in most model organisms7. Non-coding transcription is more prominent in higher eukaryotes, indicating that higher genome complexity made it necessary to make use of an additional layer of gene regulation. It should be noted that a recent yeast study showed that some antisense transcription might be an artefact arising from spurious synthesis of second-strand cDNA during reverse transcription reactions8. This indicates the amount of antisense non-coding transcription identified using strategies that did not include Actinomycin D may have been overestimated.
Classes of non-coding RNAs
Eukaryotic ncRNAs can be divided into two major classes according to their function: ncRNAs involved in processing and translating mRNAs, and, ncRNAs that play a role in regulatory role in gene expression9. The former include ribosomal RNAs (rRNAs), transfer RNAs (tRNAs), small nuclear RNAs (snRNAs) and small nucleolar RNAs (snoRNA). The latter include small (micro) and macro ncRNAs6.
Although small ncRNAs have received most attention in recent years, it is now clear that regulatory ncRNAs can be divided in two groups according to their size: ‘small’ ncRNAs and ‘macro’ ncRNAs. Small ncRNAs like miRNAs, siRNAs and rasiRNAs are transcribed as long precursor RNAs, however, only the product which is processed to lengths of 20-25bp is stably accumulated in the cell. Macro ncRNAs, in contrast are stably accumulated in the cell as very long intronless (or intron poor) transcripts that can be more than 100kb in length. Small ncRNAs have now been shown to play multiple roles in gene regulation. In mammals this regulation is predominantly at a post-transcriptional level, whereby miRNAs block the translation of mRNAs at the ribosome. In other organisms such as the yeast S. pombe or the plant A. thaliana, small ncRNAs play a role in transcriptional gene silencing and in the establishment of centromeric heterochromatin10,11.While it is not yet clear if small ncRNAs can lead to transcriptional silencing in mammals, macro ncRNAs have been clearly demonstrated to cause transcriptional gene silencing12,13. These regulatory macro ncRNAs are the focus of this review.
Macro ncRNAs and their significance in gene regulation
Macro ncRNAs have been suggested to be involved in gene regulation, sometimes only based on their negative correlation with flanking genes, for example the TK (thymidine kinase) gene14, the Keratin 1815 and the human PCNA (Proliferating Cell Nuclear Antigen) gene16. Experimental data provided stronger support that some genes are regulated by macro ncRNAs, like eIF-2a (Eukaryotic Initiation Factor 2a)17, HFE (Hereditary haemochromatosis protein precursor)18, human ß-globin19-21 and the regulation of Polycomb group response elements22.
Despite their abundance and predicted significance, the appreciation that macro ncRNAs regulate gene expression is relatively recent and little is known of how they silence genes. In most of the above examples two main models are suggested: first, a transcript-based mechanism in which the ncRNA product plays a role in silencing in cis or in trans. Second, a transcription-based mechanism in which ncRNA transcription is sufficient for silencing flanking genes in cis. The SINE encoded B2 ncRNA which binds to RNAPII and changes its transcription properties and the HOTAIR ncRNA which represses a HOX gene cluster by interaction with PRC2 (Polycomb Repressive Complex 2), are both examples whereby the macro ncRNA product plays a role in silencing23-24. The silencing of the human alpha globin gene by an upstream juxtaposed transcript in a novel type of anemia, is an example whereby the transcription of the macro ncRNA is sufficient for silencing25,26. Detailed studies of gene loci regulated by macro ncRNAs are however, rare. In contrast, a large amount of data describing gene silencing by macro ncRNAs linked to X chromosome inactivation and imprinted gene silencing is available and these studies may provide a molecular model to increase our understanding of how macro ncRNAs induce transcriptional gene silencing in the mammalian genome.
Genomic imprinting as a model for macro ncRNA mediated gene silencing
Genomic imprinting is an epigenetic phenomenon by which just one of the two parental alleles of a diploid cell is expressed in a parental specific manner. A gene that shows imprinted expression is expressed from one parental copy, for example the maternal one, and silent on the other, for example the paternal one. Imprinted genes are now known to be contained in gene clusters that are co-ordinately regulated by one imprint control element. Each imprinted gene cluster contains at least one macro ncRNA: Air in the Igf2r cluster, H19 in the Igf2 cluster, Kcnq1ot1 in the Kcnq1 gene cluster, Nespas in the Gnas cluster, Ube3aAS in the PWS/AS cluster and Gtl2 in the Dlk1 cluster27,28. While much correlative data supports a role for the macro ncRNA in regulating imprinted gene expression of the whole cluster, to date, only three imprinted macro ncRNAs have been directly tested. It has been shown that two macro ncRNAs, Air and Kcnq1ot1 are able to silence in cis multiple genes in their respective clusters29,30, while a third one, H19, has no role31. Despite the fact that expression of the H19 ncRNA negatively correlates with expression of flanking imprinted genes, imprinting control at the Igf2 cluster is mediated by the insulator function of a DMR (differentially methylated region), that lies upstream to the H19 ncRNA promoter32. The mode of action of this insulator is shown in Figure 1A.
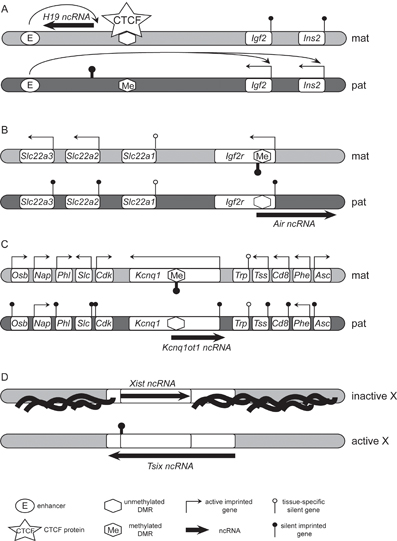

Figure 1: Macro RNAs implicated in genomic imprinting and X-inactivation
A. The Igf2 imprinted gene cluster. On the maternal chromosome (light gray) the unmethylated DMR binds the CTCF insulator protein and restricts action of enhancers to the H19 promoter resulting in maternal expression of the ncRNA. On the paternal chromosome (dark gray) the methylated DMR cannot bind CTCF and the insulator does not form resulting in paternal expression of Igf2 and Ins2. B. The Igf2r imprinted gene cluster. On the maternal chromosome (light gray) the Air ncRNA promoter is inactivated by DNA methylation of the DMR2 leading to the maternal expression of Igf2r, Slc22a2 and Slc22a3. On the paternal chromosome (dark gray) the unmethylated DMR2 allows expression of Air ncRNA that results in silencing of three protein-coding genes in cis contained in a 400kb region C. The Kcnq1 imprinted gene cluster. Methylation of the DMR on the maternal chromosome (light gray) prevents expression of the Kcnq1ot1 ncRNA and allows expression of protein coding genes Osb, Nap, Phl, Slc, Cdk, Kcnq1, Tss, Cd8 and Asc. Expression of the Kcnq1ot1 ncRNA from the paternal chromosome (dark gray) results in cis-silencing of 8 protein-coding genes spread over a 1Mbp region. The names of the genes are abbreviated as follows: Asc-Ascl2; Cd8-Cd81; Cdk-Cdkn1c; Nap-Nap1/4; Osb-Osbpl5; Phe-Phemx; Phl-Phlda2; Slc-Slc22a18; Trp-Trpm5; Tss-Tssc4. D. The X chromosomes in female cells. Antisense expression of the Tsix ncRNA prevents accumulation of the Xist ncRNA before and during the initiation of the X-inactivation. Coating and spreading of Xist on the future inactive X chromosome is accompanied by numerous epigenetic changes and leads to transcriptional silencing of the entire X chromosome. The Xist promoter is silenced by a DNA methylation mark on the active X chromosome. Note that X chromosome inactivation is random and not parental specific in somatic tissues. Adapted from Pauler et al. 2007[13].
Macro ncRNAs silence genes in cis in the Igf2r and Kcnq1 imprinted gene clusters
The imprinted Igf2r gene cluster is located on mouse chromosome 17 and spans about 400kb. The cluster harbours three maternally expressed protein-coding genes Igf2r, Slc22a2 and Slc22a3 and the paternally expressed Air ncRNA (Figure 1B). Igf2r encodes a protein, which, among other functions, acts as an essential embryonic growth suppressor while Slc22a2 and Slc22a3, encode members of the solute cation carrier family 22a2 that are not essential for development33,34. Figure 1B shows that Air ncRNA promoter is located in an antisense orientation in an Igf2r intron and overlaps the 5′ part of the Igf2r gene. Air is a 108kb long, repeat-rich, polyadenylated RNA that is transcribed by RNA Polymerase II (RNAPII)35,36. Air mainly exists as an unspliced and (relatively) unstable transcript that is retained in the nucleus at the site of transcription37. Interestingly, the silencing of the protein-coding genes in this cluster is not ubiquitous since, although Igf2r shows widespread imprinted expression in embryo, placental and adult tissues, it shows biallelic expression in post mitotic neurons. The Slc22a2 and Slc22a3 genes only show imprinted expression in placental tissues, they are biallelically expressed in adult tissues that retain imprinted expression of Igf2r and Air38,39.
The imprint that regulates imprinted expression of the whole Igf2r cluster is a DNA methylation mark on a differentially methylated region (DMR) called DMR2. DMR2 is located in the second intron of Igf2r and contains a CpG island and the Air promoter. The methylation of DMR2 is acquired in the oocyte and transmitted to the embryo where it resists the wave of demethylation that occurs in the pre-implantation embryo40. This maternal-specific methylation of DMR2 is then retained in somatic tissues, but is ultimately erased in primordial germ cells of the offspring. DMR2 is unmethylated on the paternal chromosome resulting in paternal expression of the macro Air ncRNA, while maternal methylation of DMR2 silences the maternal Air promoter36. It has been shown that truncation of the Air ncRNA from 108kb to 3kb results in loss of silencing and biallelic expression of Igf2r, Slc22a2 and Slc22a329. This data clearly demonstrates that paternal expression of the macro Air ncRNA causes bidirectional silencing in cis, of three protein coding genes spanning 400kb. Notably, Air overlaps the promoter of only one of the genes that is silenced.
The function of the paternally-expressed Kcnq1ot1 ncRNA in the Kcnq1 imprinted gene cluster (Figure 1C) resembles that of Air. Kcnq1ot1 is a paternally-expressed ncRNA whose promoter resides in an antisense orientation in a Kcnq1 intron, however, its exact length is unknown. The Kcnq1ot1 ncRNA promoter is also contained in a maternal DMR and its deletion results in loss of expression of the Kcnq1ot1 ncRNA, and loss of paternal-specific silencing at the Kcnq1 locus41. Experiments in an ex vivo episome system indicated the Kcnq1ot1 transcript is crucial for bi-directional silencing of reporter genes in cis42. The data have been confirmed in mice where truncation of the Kcnq1ot1 ncRNA to 1.5kb resulted in derepression of all 8 paternally-silenced genes in this imprinted cluster30.
The Kcnq1 and the Igf2r imprinted gene clusters are therefore regulated by the expression of a macro ncRNA. There are two main models explaining how Air or Kcnq1ot1 could exert paternal-specific silencing in their gene clusters. The RNA-directed targeting (RDT) model assumes a role for the ncRNA transcript itself, whereas the Transcription Interference (TI) model focuses on the role for transcription alone. These two models are discussed below with respect to the Igf2r imprinted gene cluster.
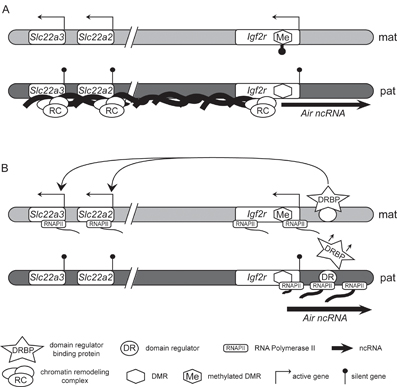

Figure 2: Two cis-acting silencing models at the Igf2r imprinted gene cluster.
A. RNA directed silencing model. On the maternal chromosome (light gray) DNA methylation of the DMR and the Air promoter results in expression of Igf2r, Slc22a2 and Slc22a3. On the paternal chromosome (dark gray) the Air ncRNA is expressed and spreads in cis over a 400kb region containing the silenced genes. The ncRNA recruits chromatin remodeling complexes resulting in silencing of the three protein-coding genes in cis. B. Transcriptional Interference (TI) model. On the maternal chromosome (light gray) RNAPII binds to the Igf2r promoter inducing Igf2r expression. Additionally, the absence of Air transcription enables the access of putative, placenta-specific domain regulator binding protein (DRBP) to the domain regulator element (DR) resulting in expression of the Slc22a2 and Slc22a3 genes. On the paternal chromosome (dark gray), transcription of the Air ncRNA in the antisense orientation interferes with the binding of RNAPII to the Igf2r promoter and also interferes with binding of the DRBP to the DR thus blocking activation of the placenta-specific domain regulator, resulting in silencing of Igf2r, Slc22a2 and Slc22a3 in cis.
A role for the ncRNA transcript?
The Air RNA directed targeting (RDT) model is based on similarities between the ncRNAs Air and Xist. Xist is a cis-acting 17kb long intron poor ncRNA involved in mammalian X-chromosome inactivation (XCI), the process of transcriptional silencing of one of the two X chromosomes in female (genotype XX) somatic cells43. Silencing starts with spreading of the Xist ncRNA on the X chromosome chosen for inactivation (Figure 1D). Note that Xist is itself regulated by an overlapping antisense ncRNA named Tsix. The up-regulation of Xist on the future inactive X chromosome is accompanied by the RNA coating of the whole chromosome that is necessary to trigger chromatin remodeling involving histone H3K27 and H3K9 hypermethylation, H3 and H4 hypoacetylation, H3K4 hypomethylation, incorporation of histone variants and DNA methylation. These multiple successive layers of modifications result in the establishment of a silent chromatin structure on the inactive X chromosome43. Air could act in a similar manner to Xist (Figure 2A). Expression of Air from the paternal allele could lead to its accumulation and limited spreading in cis followed by the recruitment of chromatin remodeling complexes, resulting in transcriptional repression of Igf2r, Slc22a2 and Slc22a3. The fact that both Xist and Air are localised in the nucleus at the site of transcription supports the RDT model, however the Xist ncRNA is more stable than Air: half-life of Xist is around 3.5-5h, whereas half-life of Air is around 1.6h36. Additionally, the applicability of an Air RNA directed targeting model is challenged by the restricted imprinted expression of Slc22a2 and Slc22a3 as these genes, in contrast to Igf2r, lack imprinted expression in adult tissues. Transcriptional analysis of X-linked genes in 8- to 16-cell embryo revealed a gradient of silencing with inverse correlation between the degree of silencing and distance from the X-inactivation center (Xic)44. A similar gradient of silencing could explain the extensive repression of Igf2r lying in the proximity of Air and less stringent imprinted silencing of Slc22a2 and Slc22a3 genes lying further. An RNA directed targeting mechanism would also imply that heterochromatic marks encompass the whole Igf2r imprinted cluster. However, a detailed tiling array analysis of the Igf2r imprinted cluster has shown that histone modifications associated with silent genes, map only to promoter regions45. These data: the relative instability of Air and the absence of spreading repressive chromatin marks, argues against an RNA directed targeting model.
A role for transcription?
It has been suggested that the Air ncRNA transcript itself has no role in the silencing process13,36. Instead, the act of Air transcription could lead to paternal repression of Igf2r, Slc22a2 and Slc22a3 by a phenomenon called Transcriptional Interference. Transcriptional Interference is a known, but rarely studied regulatory mechanism in eukaryotic genomes46. According to this model, silencing of Igf2r depends only on RNAPII continually traversing its promoter. If Air expression is lost or does not go through the Igf2r promoter, like in the case of Air truncated to 3kb, Igf2r is biallelically expressed (Figure 2B). While the instability of the mature Air ncRNA indicates some support for a Transcription Interference model where only transcription is necessary, the nuclear retention of the transcript does not. However, nuclear retention might only be the result of impaired splicing of the Air ncRNA36.
Once again interesting parallels between X-inactivation and imprinting at the Igf2r locus can be drawn. Tsix is a macro ncRNA, antisense to Xist that regulates the X-inactivation process by silencing Xist (Figure 1D). Tsix and Air share some similarities: both are repeat-rich unspliced non-coding ncRNAs expressed in an antisense orientation, both overlap the promoters of their counterpart sense genes and regulate their expression. Truncation of Tsix results in accumulation of Xist from the modified chromosome47. Similarly, truncation of Air to 3kb results in up-regulated expression of Igf2r on the truncated allele. It has been suggested that Tsix silences Xist by a transcription based mechanism48 and it has been shown recently that Tsix transcription has to traverse through the Xist promoter to silence Xist49. However, one obvious difference exists between Tsix and Air. The silencing function of Tsix is only targeted towards one gene, Xist. Therefore, an additional mechanism must be involved on chromosome 17, which keeps both Slc22a2 and Slc22a3 silent on the paternal allele.
Since transcription of Air does not overlap Slc22a2 and Slc22a3, it is difficult to imagine how these genes should be silenced by Transcriptional Interference. However, a model has been suggested by which it can be explained36. In this model it is proposed that in early placenta, expression of Slc22a2 and Slc22a3 is dependent on activity of a domain regulator (for example, placental enhancer) contained within the Air gene body (Figure 2B). The paternal domain regulator is, however, disturbed by Air transcription. In other tissues of the embryo and the adult mouse expression of these genes is independent of this domain regulator, for example because other activator elements take over and Slc22a2 and Slc22a3 become biallelically expressed. A placental-specific domain regulator for Slc22a2 and Slc22a3 lying inside the 108kb long Air gene body has the potential to explain both, the imprinted expression of these two genes and tissue specificity of this imprinted expression. However to date this putative domain regulator has not been identified.
Implications of macro ncRNAs for epigenetic therapy
In mammals, macro ncRNAs are regulators of gene expression on the inactive X chromosome, in imprinted gene clusters and at some non-imprinted loci. Therefore misregulation of macro ncRNAs could potentially result in impaired expression of flanking protein coding genes. It is important to note that imprinted macro ncRNAs such as Air and Kcnq1ot1 as well as the Xist ncRNA, are directly silenced by DNA methylation and thus become expressed from both parental chromosomes in the absence of genome-wide DNA methylation. As a consequence of macro ncRNA deregulation, the flanking protein-coding genes become silenced on both parental chromosomes36,50-51. This indicates that epigenetic therapies based on demethylating agents such Decitabine, could change macro ncRNA expression particularly at imprinted gene clusters and on the inactive X chromosome. Since many of the imprinted protein-coding genes that are silenced by macro ncRNAs are growth suppressors (for example, Igf2r and Cdkn1), this would not be a desirable goal for epigenetic therapy. The use of genomic imprinting as a model of epigenetic gene silencing by macro ncRNAs can help us to understand how genes are regulated and, as a final goal, to develop safe epigenetic therapies.
References
- Carninci, P., et al., The transcriptional landscape of the mammalian genome. Science, 2005. 309(5740): p. 1559-63.
- Cawley, S., et al., Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell, 2004. 116(4): p. 499-509.
- Cheng, J., et al., Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science, 2005. 308(5725):p. 1149-54.
- Carninci, P., et al., Targeting a complex transcriptome: the construction of the mouse full-length cDNA encyclopedia. Genome Res, 2003. 13(6B): p. 1273-89.
- Kapranov, P., et al., RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science, 2007. 316(5830): p. 1484-8.
- Mattick, J.S., The functional genomics of noncoding RNA. Science, 2005. 309(5740): p. 1527-8.
- Shabalina, S.A. and N.A. Spiridonov, The mammalian transcriptome and the function of non-coding DNA sequences. Genome Biol, 2004. 5(4): p. 105.
- Perocchi, F., et al., Antisense artifacts in transcriptome microarray experiments are resolved by actinomycin D. Nucleic Acids Res, 2007. 35(19): p. e128.
- Morey, C. and P. Avner, Employment opportunities for non-coding RNAs. FEBS Lett, 2004. 567(1): p. 27-34.
- Kanellopoulou, C., et al., Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev, 2005. 19(4): p. 489-501.
- Zaratiegui, M., D.V. Irvine, and R.A. Martienssen, Noncoding RNAs and gene silencing. Cell, 2007. 128(4): p. 763-76.
- Prasanth, K.V. and D.L. Spector, Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum. Genes Dev, 2007. 21(1): p. 11-42.
- Pauler, F.M., M.V. Koerner, and D.P. Barlow, Silencing by imprinted noncoding RNAs: is transcription the answer? Trends Genet, 2007.
- Sutterluety, H., et al., Growth-regulated antisense transcription of the mouse thymidine kinase gene. Nucleic Acids Res, 1998. 26(21): p. 4989-95.
- Neznanov, N.S. and R.G. Oshima, cis regulation of the keratin 18 gene in transgenic mice. Mol Cell Biol, 1993. 13(3): p. 1815-23.
- Tommasi, S. and G.P. Pfeifer, In vivo structure of two divergent promoters at the human PCNA locus. Synthesis of antisense RNA and S phase-dependent binding of E2F complexes in intron 1. J Biol Chem, 1999. 274(39): p. 27829-38.
- Noguchi, M., et al., Characterization of an antisense Inr element in the eIF-2 alpha gene. J Biol Chem, 1994. 269(46): p. 29161-7.
- Thenie, A.C., et al., Identification of an endogenous RNA transcribed from the antisense strand of the HFE gene. Hum Mol Genet, 2001. 10(17): p. 1859-66.
- Plant, K.E., S.J. Routledge, and N.J. Proudfoot, Intergenic transcription in the human beta-globin gene cluster. Mol Cell Biol, 2001. 21(19): p. 6507-14.
- Gribnau, J., et al., Intergenic transcription and developmental remodeling of chromatin subdomains in the human beta-globin locus. Mol Cell, 2000. 5(2): p. 377-86.
- Routledge, S.J. and N.J. Proudfoot, Definition of transcriptional promoters in the human beta globin locus control region. J Mol Biol, 2002. 323(4): p. 601-11.
- Schmitt, S., M. Prestel, and R. Paro, Intergenic transcription through a polycomb group response element counteracts silencing. Genes Dev, 2005. 19(6): p. 697-708.
- Espinoza, C.A., et al., B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat Struct Mol Biol, 2004. 11(9): p. 822-9.
- Rinn, J.L., et al., Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell, 2007. 129(7): p. 1311-23.
- Martianov, I., et al., Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature, 2007. 445(7128): p. 666-70.
- Tufarelli, C., et al., Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat Genet, 2003. 34(2): p. 157-65.
- Wood, A.J. and R.J. Oakey, Genomic imprinting in mammals: emerging themes and established theories. PLoS Genet, 2006. 2(11): p. e147.
- Barlow, D.P. and M.S. Bartolomei, Genomic Imprinting in Mammals, in Epigenetics, C.D. Allis, T. Jenuwein, and D. Reinberg, Editors. 2006, Cold Spring Harbor Labroratory Press: Cold Spring Harbor, New York. p. 357-377.
- Sleutels, F., R. Zwart, and D.P. Barlow, The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature, 2002. 415(6873): p. 810-3.
- Mancini-Dinardo, D., et al., Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev, 2006. 20(10): p. 1268-82.
- Jones, B.K., J.M. Levorse, and S.M. Tilghman, Igf2 imprinting does not require its own DNA methylation or H19 RNA. Genes Dev, 1998. 12(14): p. 2200-7.
- Lewis, A. and A. Murrell, Genomic imprinting: CTCF protects the boundaries. Curr Biol, 2004. 14(7): p. R284-6.
- Wang, Z.Q., et al., Regulation of embryonic growth and lysosomal targeting by the imprinted Igf2/Mpr gene. Nature, 1994. 372(6505): p. 464-7.
- Jonker, J.W. and A.H. Schinkel, Pharmacological and physiological functions of the polyspecific organic cation transporters: OCT1, 2, and 3 (SLC22A1-3). J Pharmacol Exp Ther, 2004. 308(1): p. 2-9.
- Lyle, R., et al., The imprinted antisense RNA at the Igf2r locus overlaps but does not imprint Mas1. Nat Genet, 2000. 25(1): p. 19-21.
- Seidl, C.I., S.H. Stricker, and D.P. Barlow, The imprinted Air ncRNA is an atypical RNAPII transcript that evades splicing and escapes nuclear export. Embo J, 2006. 25(15): p. 3565-75.
- Braidotti, G., et al., The Air noncoding RNA: an imprinted cis-silencing transcript. Cold Spring Harb Symp Quant Biol, 2004. 69: p. 55-66.
- Yamasaki, Y., et al., Neuron-specific relaxation of Igf2r imprinting is associated with neuron-specific histone modifications and lack of its antisense transcript Air. Hum Mol Genet, 2005. 14(17): p. 2511-20.
- Zwart, R., et al., Bidirectional action of the Igf2r imprint control element on upstream and downstream imprinted genes. Genes Dev, 2001. 15(18): p. 2361-6.
- Stoger, R., et al., Maternal-specific methylation of the imprinted mouse Igf2r locus identifies the expressed locus as carrying the imprinting signal. Cell, 1993. 73(1): p. 61-71.
- Fitzpatrick, G.V., P.D. Soloway, and M.J. Higgins, Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat Genet, 2002. 32(3): p. 426-31.
- Thakur, N., et al., An antisense RNA regulates the bidirectional silencing property of the Kcnq1 imprinting control region. Mol Cell Biol, 2004. 24(18): p. 7855-62.
- Cohen, H.R., et al., Chromatin modifications on the inactive X chromosome. Prog Mol Subcell Biol, 2005. 38: p. 91-122.
- Huynh, K.D. and J.T. Lee, Inheritance of a pre-inactivated paternal X chromosome in early mouse embryos. Nature, 2003. 426(6968): p. 857-62.
- Regha, K., et al., Active and repressive chromatin are interspersed without spreading in an imprinted gene cluster in the mammalian genome. Mol Cell, 2007. 27(3): p. 353-66.
- Shearwin, K.E., B.P. Callen, and J.B. Egan, Transcriptional interference–a crash course. Trends Genet, 2005. 21(6): p. 339-45.
- Luikenhuis, S., A. Wutz, and R. Jaenisch, Antisense transcription through the Xist locus mediates Tsix function in embryonic stem cells. Mol Cell Biol, 2001. 21(24): p. 8512-20.
- Sun, B.K., A.M. Deaton, and J.T. Lee, A Transient Heterochromatic State in Xist Preempts X Inactivation Choice without RNA Stabilization. Mol Cell, 2006. 21(5): p. 617-628.
- Ohhata, T., et al., Crucial role of antisense transcription across the Xist promoter in Tsix-mediated Xist chromatin modification. Development, 2008. 135(2): p. 227-35.
- Li, E., C. Beard, and R. Jaenisch, Role for DNA methylation in genomic imprinting. Nature, 1993. 366(6453): p. 362-5.
- Beard, C., E. Li, and R. Jaenisch, Loss of methylation activates Xist in somatic but not in embryonic cells. Genes Dev, 1995. 9(19): p. 2325-34.
Denise P. Barlow
Denise is a senior scientist at the Research Center for Molecular Medicine of the Austrian Academy of Science in Vienna, Austria. She received her Ph.D. in 1981 in the lab of Derek Burke at Warwick University UK, and then undertook Post Doctoral research in the lab of Brigid Hogan at ICRF, London UK and Hans Lehrach at EMBL, Heidelberg. Her research group at the IMP Vienna discovered the first imprinted gene in 1991 and since then her research focus has been to understand the molecular basis of epigenetic silencing of imprinted genes in mammals.
Stefan H. Stricker and Paulina Latos
Stefan and Paulina are both senior PhD students in the Barlow lab where they study features of Air mediated gene silencing in an ES cell model system they helped to develop. Stefan received his Diploma in Biology from the Ludwig Maximilian University in Munich, Germany, working in the lab of Prof. Dr. Magdalena Götz on neurogenesis in the developing mouse brain. Paulina received her Master’s Degree in Biotechnology from Adam Mickiewicz University, Poznan, Poland, working in the lab of Prof. Jan Barciszewski on epigenetic aspects of hybridogenesis in water frogs.




