Learning from clinical microbiologists: culturomics and metagenomics
Posted: 20 December 2019 | Jeanne Moldenhauer (Excellent Pharma Consulting) | No comments yet
Clinical microbiologists have started using new, non-conventional methods to study the microbiology of the human gut. In this article, Jeanne Moldenhauer discusses the intricacies of culturomic and metagenomic studies and how this can advance current research.

Introduction
In recent years there has been significant interest in the sequencing of the human microbiome and what can be learned from this project. One of the areas of interest includes those normally occurring microorganisms in the human gut. It is believed that the human gut microbiota is responsible for maintaining health as well as disease pathogenesis.1 However, study of the gut microbiota has required the development of new culture and evaluation methods. Research has indicated that as many as 80 percent of the human gut bacteria are unknown and considered “unculturable”.1 This is similar to the term used by many pharmaceutical microbiologists – viable but not culturable (VBNC). To remedy this, clinical microbiologists started using new, non-conventional methods; referred to as the “rebirth of culture in microbiology”.1
Metagenomics
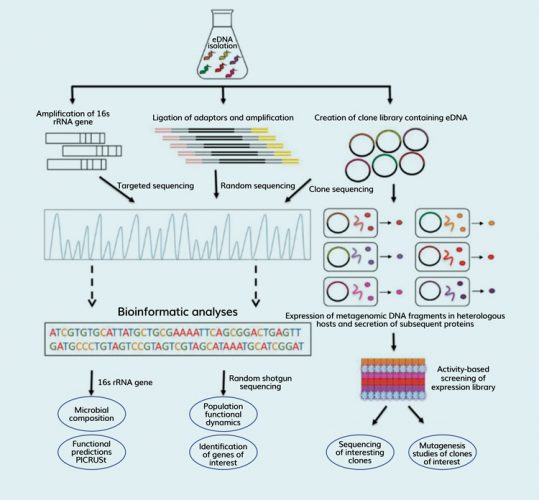

Figure 1: Metagenomics approach
Metagenomics is a term that refers to the study of genetic material that comes directly from environmental samples, rather than pure colonies of organisms. This is followed by sequencing, depicted in Figure 1, which allows for the detection of a largely unbiased sample of all genes from the sampled communities (ie, they may not be pure cultures).2 The sequencing process in metagenomics includes use of shotgun or PCR-directed sequencing. Shotgun sequencing involves sampling environmental growth from its natural habitat, filtering the particles, lysis and DNA extraction, construction of the cloning library, sequencing of the clones and assembly of the sequence into contigs and scaffolds. This flow is depicted in Figure 2.2
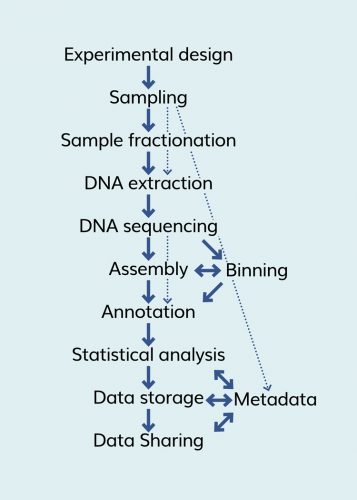

Figure 2: Shotgun metagenome process flow2
Metagenomic studies have been useful in recent years, due to their ability to assess microbial composition relatively quickly using high-throughput sequencing. Low concentration ecosystems have not been detected in some of these systems (<104–105 cells/mL). Metagenomics has replaced molecular techniques based upon PCR amplification steps, since these techniques were unable to provide information on the microbiota, only on eubacteria. Microarrays were also historically used but were not found as useful for the microbiota. Another concern with using microarrays is that they only identify known bacterial species located on the chip.3
While there are many uses for metagenomics, this technology also has limitations. One disadvantage is that the result provides only a partial snapshot of the microbiota in the human gut. A partial result can be retrieved using light microscopy observations; looking for disparity between the proportions of Gram-positive and Gram-negative organisms. A partial result can also be retrieved using pyrosequencing. Another disadvantage or limitation is that the DNA extraction process has bias.4
The extraction protocols used have also been shown to influence the final bacterial taxonomic distribution.5 It has been reported that the DNA extraction process yields the greatest number of variations,4 while other concerns relate to determination of the activity of each type of bacterium, since metagenomics cannot identify if the sequences belong to active or quiescent microorganisms. However, this may be mitigated using RNA sequencing (also known as metatranscriptomics). Another issue is the interpretation of the sequences for unknown species, since most of the gut microorganisms have not been cultured.4
Culturomics
Culturomics is a newer approach that utilises “multiple culture conditions combined with prolonged incubation”.1 This approach has resulted in the isolation of hundreds of new bacterial species from the gut in a short timeframe.1 Once isolated, sequencing is used to identify the organisms or identify that a new species exists and needs to be classified.
The benefits of culturomics include: the ability to detect minority populations (low level concentrations); the methods are not restricted to the recovery of eubacteria; and that strains are provided and can be extensively characterised.3 An example of a culturomics process is depicted in Figure 3.
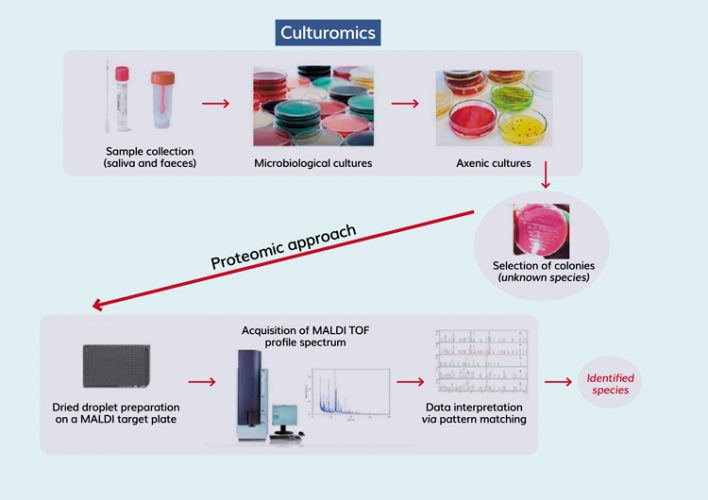

Figure 3: Culturomics approach
Using this approach, a report found that as many as 32,500 different colonies could be recovered from just three human stool samples.1 However, the ability to process all these samples is only possible with high-throughput systems like MALDI-TOF MS. Some scientists may also couple these technologies with smart incubators and automated colony-picking systems.3
The research organisation Soft Cell Biological Research has developed culture techniques for blood and urinary tract infections.6 To date, these techniques have resulted in the following: discovery of 191 new bacterial species; discovery of 696 new bacterial subspecies; and discovery of 105 known L-form bacteria. As such, these newer techniques are not limited to only gut microbiota.
Comparing metagenomics and culturomics
Table 1 compares the two different processes.
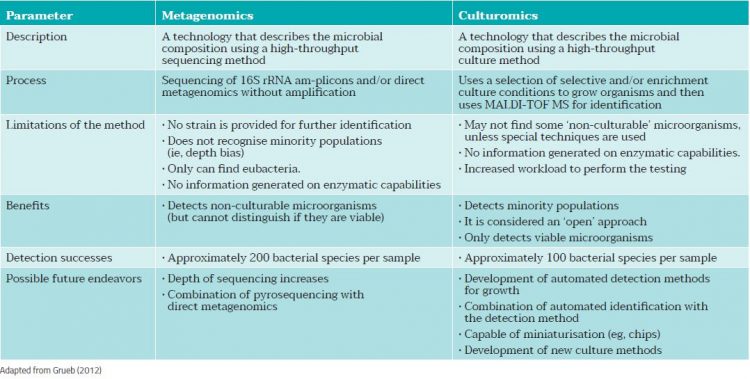

Table 1: Comparing Metagenomics and Culturomics3
Development of new culture methodologies
As a result of culturomics, many new culture methods have been developed. Examples of these methods developed for the gut microbiota include:
Dilution cultures
This technique involves the use of successive dilutions of the microbial population, continuing to a point where no bacterial growth is detected.1
Use of sea water components
For culture of Candidatus pelagibacter ubique (also known as SAR 11), which had been previously non-culturable. Addition of sea water components to the media allowed for successful culturing.1
Use of diffusion chambers
This method utilises diffusion chambers with two membranes. One membrane allows the environmental nutrients to enter the chamber. The second membrane confines the bacterial cells. In effect this simulates actual environmental conditions.1
Recovery and culture of obligate anaerobes
Several different media have been developed. One medium was supplemented with carbon sources, macroelements, metals and some growth factors. Another medium was based upon Schaedler agar supplemented with high doses of glutathione and ascorbic acid. Using the modified Schaedler agar, obligate anaerobes were able to be successfully cultured under aerobic conditions. It is believed that the high level of antioxidants (compared to earlier studies) may play a role. Another larger study was conducted adding uric acid to the culture medium. In this study 276 bacterial species, of which 82 were obligate parasites, were able to be cultured aerobically.1
Conclusion
As we learn more about the human microbiome, we have become aware of the need to develop new or modified methods to culture and identify these organisms. The data developed to date indicates many more species and subspecies of microorganisms that have not been previously discovered. While many microbiologists are focused on rapid or alternative methods for detection, enumeration and identification, there is also a space for the necessary development of new and innovation culture methods to further our knowledge. Clinical microbiologists are developing these methods and then identifying the microorganisms with newer identification methods such as MALDI-TOF.
About the author
Jeanne Moldenhauer is Vice President of Excellent Pharma Consulting and has more than 30 years’ experience in the pharmaceutical industry. She chaired the Environmental Monitoring/Microbiology Interest Group of the PDA for more than 15 years, served on the Scientific Advisory Board of the PDA for 20 years, founded the Rapid Microbiology User’s GroupTM and is a member of ASQ and RAPS.
References
- Lagier J-C, Dubourg G, Million M, Cadoret MB, Fenollar F, Levasseur A, et al. Culturing the human microbiota and culturomics. Nature Micro Reviews. 2018;16:540 – 550.
- Thomas T, Gilbert J, Meyer F. Metagenomics – a guide from sampling to data analysis. Microbial Informatics and Experimentation. 2012;2(1):3. Accessed from Wikipedia 5th September 2018.
- Greub G. Culturomics: a new approach to study the human microbiome. Clinical Microbiology and Infection. European Society of Clinical Microbiology and Infectious Diseases. 2012;18(12):1157-1159.
- Amrane S, Raoult D, Lagier J-C. Metagenomics, culturomics, and the human gut microbiota. Expert Review of Anti-infective Therapy .2018;16:5:373-375. Accessed from: https://doi.org/10.1080/14787210.2018.1467268 on 5thSeptember 2018.
- Angelakis E, Bachar D, Henrissat B, et al. Glycans affect DNA extraction and induce substantial differences in gut metagenomic studies. Sci Rep. 2016;18(6):26276.
- Soft Cell Biological Research. Our facts. Soft Cell Bio. Accessed from http://www.softcellbio.com on 9th September 2018.
Issue
Related topics
culturomics, metagenomics, Microarrays, Microbial Detection, Microbiomes
Related organisations
Agilent Technologies, Hyglos-bioMerieux, Soft Cell Biological Research, WITec





